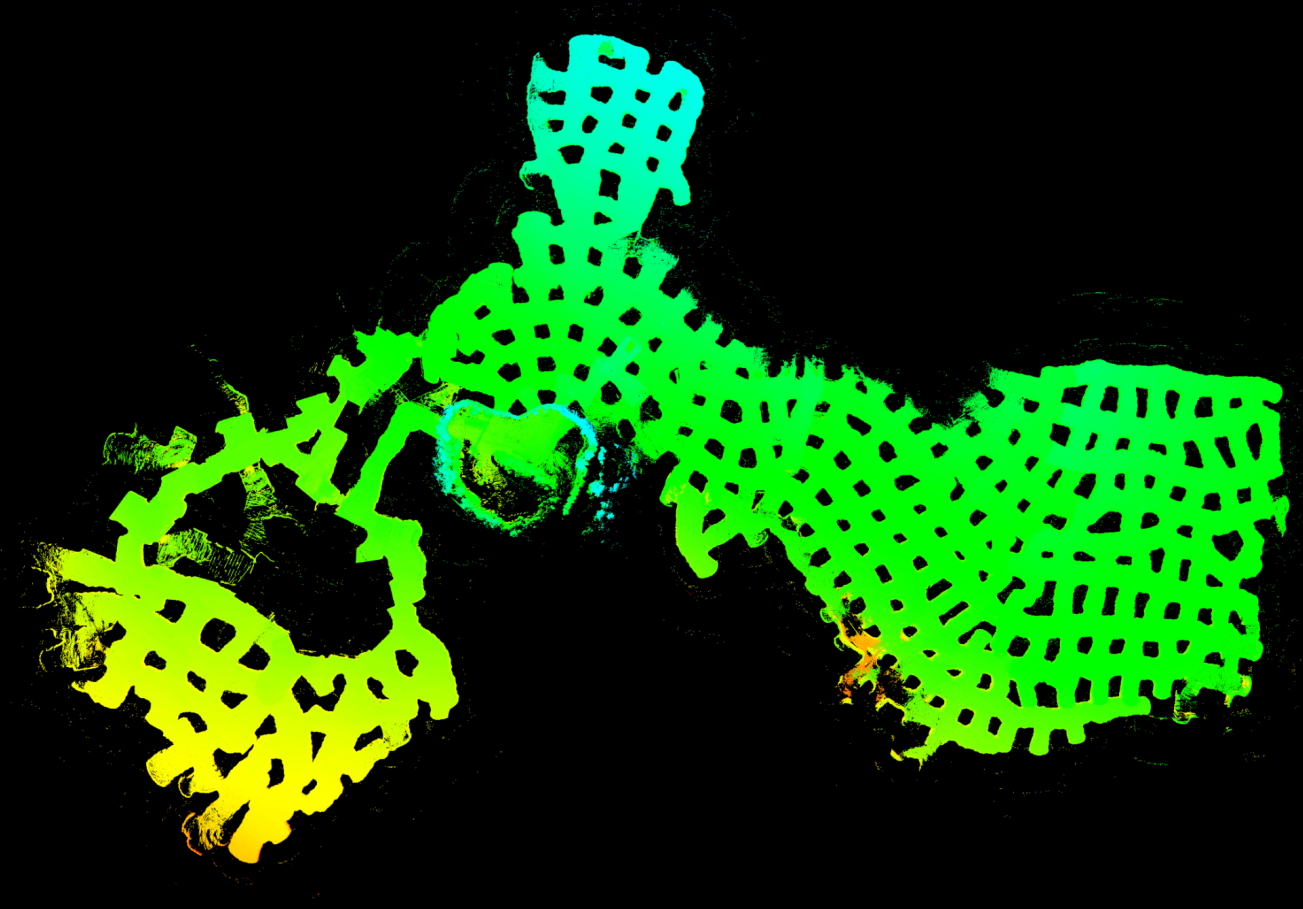
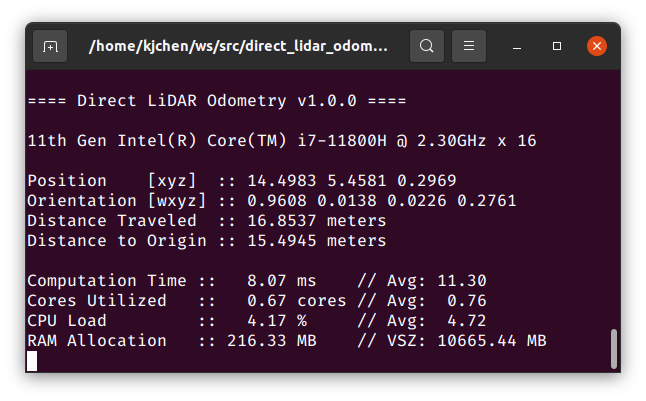
|
|
@@ -0,0 +1,2057 @@
|
|
|
+/************************************************************
|
|
|
+ *
|
|
|
+ * Copyright (c) 2021, University of California, Los Angeles
|
|
|
+ *
|
|
|
+ * Authors: Kenny J. Chen, Brett T. Lopez
|
|
|
+ * Contact: kennyjchen@ucla.edu, btlopez@ucla.edu
|
|
|
+ *
|
|
|
+ ***********************************************************/
|
|
|
+
|
|
|
+/***********************************************************************
|
|
|
+ * Software License Agreement (BSD License)
|
|
|
+ *
|
|
|
+ * Copyright 2008-2009 Marius Muja (mariusm@cs.ubc.ca). All rights reserved.
|
|
|
+ * Copyright 2008-2009 David G. Lowe (lowe@cs.ubc.ca). All rights reserved.
|
|
|
+ * Copyright 2011-2021 Jose Luis Blanco (joseluisblancoc@gmail.com).
|
|
|
+ * All rights reserved.
|
|
|
+ *
|
|
|
+ * THE BSD LICENSE
|
|
|
+ *
|
|
|
+ * Redistribution and use in source and binary forms, with or without
|
|
|
+ * modification, are permitted provided that the following conditions
|
|
|
+ * are met:
|
|
|
+ *
|
|
|
+ * 1. Redistributions of source code must retain the above copyright
|
|
|
+ * notice, this list of conditions and the following disclaimer.
|
|
|
+ * 2. Redistributions in binary form must reproduce the above copyright
|
|
|
+ * notice, this list of conditions and the following disclaimer in the
|
|
|
+ * documentation and/or other materials provided with the distribution.
|
|
|
+ *
|
|
|
+ * THIS SOFTWARE IS PROVIDED BY THE AUTHOR ``AS IS'' AND ANY EXPRESS OR
|
|
|
+ * IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED WARRANTIES
|
|
|
+ * OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE DISCLAIMED.
|
|
|
+ * IN NO EVENT SHALL THE AUTHOR BE LIABLE FOR ANY DIRECT, INDIRECT,
|
|
|
+ * INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL DAMAGES (INCLUDING, BUT
|
|
|
+ * NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR SERVICES; LOSS OF USE,
|
|
|
+ * DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER CAUSED AND ON ANY
|
|
|
+ * THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY, OR TORT
|
|
|
+ * (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE OF
|
|
|
+ * THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
|
|
|
+ *************************************************************************/
|
|
|
+
|
|
|
+/** \mainpage nanoflann C++ API documentation
|
|
|
+ * nanoflann is a C++ header-only library for building KD-Trees, mostly
|
|
|
+ * optimized for 2D or 3D point clouds.
|
|
|
+ *
|
|
|
+ * nanoflann does not require compiling or installing, just an
|
|
|
+ * #include <nanoflann.hpp> in your code.
|
|
|
+ *
|
|
|
+ * See:
|
|
|
+ * - <a href="modules.html" >C++ API organized by modules</a>
|
|
|
+ * - <a href="https://github.com/jlblancoc/nanoflann" >Online README</a>
|
|
|
+ * - <a href="http://jlblancoc.github.io/nanoflann/" >Doxygen
|
|
|
+ * documentation</a>
|
|
|
+ */
|
|
|
+
|
|
|
+#ifndef NANOFLANN_HPP_
|
|
|
+#define NANOFLANN_HPP_
|
|
|
+
|
|
|
+#include <algorithm>
|
|
|
+#include <array>
|
|
|
+#include <cassert>
|
|
|
+#include <cmath> // for abs()
|
|
|
+#include <cstdio> // for fwrite()
|
|
|
+#include <cstdlib> // for abs()
|
|
|
+#include <functional>
|
|
|
+#include <limits> // std::reference_wrapper
|
|
|
+#include <stdexcept>
|
|
|
+#include <vector>
|
|
|
+
|
|
|
+/** Library version: 0xMmP (M=Major,m=minor,P=patch) */
|
|
|
+#define NANOFLANN_VERSION 0x132
|
|
|
+
|
|
|
+// Avoid conflicting declaration of min/max macros in windows headers
|
|
|
+#if !defined(NOMINMAX) && \
|
|
|
+ (defined(_WIN32) || defined(_WIN32_) || defined(WIN32) || defined(_WIN64))
|
|
|
+#define NOMINMAX
|
|
|
+#ifdef max
|
|
|
+#undef max
|
|
|
+#undef min
|
|
|
+#endif
|
|
|
+#endif
|
|
|
+
|
|
|
+namespace nanoflann {
|
|
|
+/** @addtogroup nanoflann_grp nanoflann C++ library for ANN
|
|
|
+ * @{ */
|
|
|
+
|
|
|
+/** the PI constant (required to avoid MSVC missing symbols) */
|
|
|
+template <typename T> T pi_const() {
|
|
|
+ return static_cast<T>(3.14159265358979323846);
|
|
|
+}
|
|
|
+
|
|
|
+/**
|
|
|
+ * Traits if object is resizable and assignable (typically has a resize | assign
|
|
|
+ * method)
|
|
|
+ */
|
|
|
+template <typename T, typename = int> struct has_resize : std::false_type {};
|
|
|
+
|
|
|
+template <typename T>
|
|
|
+struct has_resize<T, decltype((void)std::declval<T>().resize(1), 0)>
|
|
|
+ : std::true_type {};
|
|
|
+
|
|
|
+template <typename T, typename = int> struct has_assign : std::false_type {};
|
|
|
+
|
|
|
+template <typename T>
|
|
|
+struct has_assign<T, decltype((void)std::declval<T>().assign(1, 0), 0)>
|
|
|
+ : std::true_type {};
|
|
|
+
|
|
|
+/**
|
|
|
+ * Free function to resize a resizable object
|
|
|
+ */
|
|
|
+template <typename Container>
|
|
|
+inline typename std::enable_if<has_resize<Container>::value, void>::type
|
|
|
+resize(Container &c, const size_t nElements) {
|
|
|
+ c.resize(nElements);
|
|
|
+}
|
|
|
+
|
|
|
+/**
|
|
|
+ * Free function that has no effects on non resizable containers (e.g.
|
|
|
+ * std::array) It raises an exception if the expected size does not match
|
|
|
+ */
|
|
|
+template <typename Container>
|
|
|
+inline typename std::enable_if<!has_resize<Container>::value, void>::type
|
|
|
+resize(Container &c, const size_t nElements) {
|
|
|
+ if (nElements != c.size())
|
|
|
+ throw std::logic_error("Try to change the size of a std::array.");
|
|
|
+}
|
|
|
+
|
|
|
+/**
|
|
|
+ * Free function to assign to a container
|
|
|
+ */
|
|
|
+template <typename Container, typename T>
|
|
|
+inline typename std::enable_if<has_assign<Container>::value, void>::type
|
|
|
+assign(Container &c, const size_t nElements, const T &value) {
|
|
|
+ c.assign(nElements, value);
|
|
|
+}
|
|
|
+
|
|
|
+/**
|
|
|
+ * Free function to assign to a std::array
|
|
|
+ */
|
|
|
+template <typename Container, typename T>
|
|
|
+inline typename std::enable_if<!has_assign<Container>::value, void>::type
|
|
|
+assign(Container &c, const size_t nElements, const T &value) {
|
|
|
+ for (size_t i = 0; i < nElements; i++)
|
|
|
+ c[i] = value;
|
|
|
+}
|
|
|
+
|
|
|
+/** @addtogroup result_sets_grp Result set classes
|
|
|
+ * @{ */
|
|
|
+template <typename _DistanceType, typename _IndexType = size_t,
|
|
|
+ typename _CountType = size_t>
|
|
|
+class KNNResultSet {
|
|
|
+public:
|
|
|
+ typedef _DistanceType DistanceType;
|
|
|
+ typedef _IndexType IndexType;
|
|
|
+ typedef _CountType CountType;
|
|
|
+
|
|
|
+private:
|
|
|
+ IndexType *indices;
|
|
|
+ DistanceType *dists;
|
|
|
+ CountType capacity;
|
|
|
+ CountType count;
|
|
|
+
|
|
|
+public:
|
|
|
+ inline KNNResultSet(CountType capacity_)
|
|
|
+ : indices(0), dists(0), capacity(capacity_), count(0) {}
|
|
|
+
|
|
|
+ inline void init(IndexType *indices_, DistanceType *dists_) {
|
|
|
+ indices = indices_;
|
|
|
+ dists = dists_;
|
|
|
+ count = 0;
|
|
|
+ if (capacity)
|
|
|
+ dists[capacity - 1] = (std::numeric_limits<DistanceType>::max)();
|
|
|
+ }
|
|
|
+
|
|
|
+ inline CountType size() const { return count; }
|
|
|
+
|
|
|
+ inline bool full() const { return count == capacity; }
|
|
|
+
|
|
|
+ /**
|
|
|
+ * Called during search to add an element matching the criteria.
|
|
|
+ * @return true if the search should be continued, false if the results are
|
|
|
+ * sufficient
|
|
|
+ */
|
|
|
+ inline bool addPoint(DistanceType dist, IndexType index) {
|
|
|
+ CountType i;
|
|
|
+ for (i = count; i > 0; --i) {
|
|
|
+#ifdef NANOFLANN_FIRST_MATCH // If defined and two points have the same
|
|
|
+ // distance, the one with the lowest-index will be
|
|
|
+ // returned first.
|
|
|
+ if ((dists[i - 1] > dist) ||
|
|
|
+ ((dist == dists[i - 1]) && (indices[i - 1] > index))) {
|
|
|
+#else
|
|
|
+ if (dists[i - 1] > dist) {
|
|
|
+#endif
|
|
|
+ if (i < capacity) {
|
|
|
+ dists[i] = dists[i - 1];
|
|
|
+ indices[i] = indices[i - 1];
|
|
|
+ }
|
|
|
+ } else
|
|
|
+ break;
|
|
|
+ }
|
|
|
+ if (i < capacity) {
|
|
|
+ dists[i] = dist;
|
|
|
+ indices[i] = index;
|
|
|
+ }
|
|
|
+ if (count < capacity)
|
|
|
+ count++;
|
|
|
+
|
|
|
+ // tell caller that the search shall continue
|
|
|
+ return true;
|
|
|
+ }
|
|
|
+
|
|
|
+ inline DistanceType worstDist() const { return dists[capacity - 1]; }
|
|
|
+};
|
|
|
+
|
|
|
+/** operator "<" for std::sort() */
|
|
|
+struct IndexDist_Sorter {
|
|
|
+ /** PairType will be typically: std::pair<IndexType,DistanceType> */
|
|
|
+ template <typename PairType>
|
|
|
+ inline bool operator()(const PairType &p1, const PairType &p2) const {
|
|
|
+ return p1.second < p2.second;
|
|
|
+ }
|
|
|
+};
|
|
|
+
|
|
|
+/**
|
|
|
+ * A result-set class used when performing a radius based search.
|
|
|
+ */
|
|
|
+template <typename _DistanceType, typename _IndexType = size_t>
|
|
|
+class RadiusResultSet {
|
|
|
+public:
|
|
|
+ typedef _DistanceType DistanceType;
|
|
|
+ typedef _IndexType IndexType;
|
|
|
+
|
|
|
+public:
|
|
|
+ const DistanceType radius;
|
|
|
+
|
|
|
+ std::vector<std::pair<IndexType, DistanceType>> &m_indices_dists;
|
|
|
+
|
|
|
+ inline RadiusResultSet(
|
|
|
+ DistanceType radius_,
|
|
|
+ std::vector<std::pair<IndexType, DistanceType>> &indices_dists)
|
|
|
+ : radius(radius_), m_indices_dists(indices_dists) {
|
|
|
+ init();
|
|
|
+ }
|
|
|
+
|
|
|
+ inline void init() { clear(); }
|
|
|
+ inline void clear() { m_indices_dists.clear(); }
|
|
|
+
|
|
|
+ inline size_t size() const { return m_indices_dists.size(); }
|
|
|
+
|
|
|
+ inline bool full() const { return true; }
|
|
|
+
|
|
|
+ /**
|
|
|
+ * Called during search to add an element matching the criteria.
|
|
|
+ * @return true if the search should be continued, false if the results are
|
|
|
+ * sufficient
|
|
|
+ */
|
|
|
+ inline bool addPoint(DistanceType dist, IndexType index) {
|
|
|
+ if (dist < radius)
|
|
|
+ m_indices_dists.push_back(std::make_pair(index, dist));
|
|
|
+ return true;
|
|
|
+ }
|
|
|
+
|
|
|
+ inline DistanceType worstDist() const { return radius; }
|
|
|
+
|
|
|
+ /**
|
|
|
+ * Find the worst result (furtherest neighbor) without copying or sorting
|
|
|
+ * Pre-conditions: size() > 0
|
|
|
+ */
|
|
|
+ std::pair<IndexType, DistanceType> worst_item() const {
|
|
|
+ if (m_indices_dists.empty())
|
|
|
+ throw std::runtime_error("Cannot invoke RadiusResultSet::worst_item() on "
|
|
|
+ "an empty list of results.");
|
|
|
+ typedef
|
|
|
+ typename std::vector<std::pair<IndexType, DistanceType>>::const_iterator
|
|
|
+ DistIt;
|
|
|
+ DistIt it = std::max_element(m_indices_dists.begin(), m_indices_dists.end(),
|
|
|
+ IndexDist_Sorter());
|
|
|
+ return *it;
|
|
|
+ }
|
|
|
+};
|
|
|
+
|
|
|
+/** @} */
|
|
|
+
|
|
|
+/** @addtogroup loadsave_grp Load/save auxiliary functions
|
|
|
+ * @{ */
|
|
|
+template <typename T>
|
|
|
+void save_value(FILE *stream, const T &value, size_t count = 1) {
|
|
|
+ fwrite(&value, sizeof(value), count, stream);
|
|
|
+}
|
|
|
+
|
|
|
+template <typename T>
|
|
|
+void save_value(FILE *stream, const std::vector<T> &value) {
|
|
|
+ size_t size = value.size();
|
|
|
+ fwrite(&size, sizeof(size_t), 1, stream);
|
|
|
+ fwrite(&value[0], sizeof(T), size, stream);
|
|
|
+}
|
|
|
+
|
|
|
+template <typename T>
|
|
|
+void load_value(FILE *stream, T &value, size_t count = 1) {
|
|
|
+ size_t read_cnt = fread(&value, sizeof(value), count, stream);
|
|
|
+ if (read_cnt != count) {
|
|
|
+ throw std::runtime_error("Cannot read from file");
|
|
|
+ }
|
|
|
+}
|
|
|
+
|
|
|
+template <typename T> void load_value(FILE *stream, std::vector<T> &value) {
|
|
|
+ size_t size;
|
|
|
+ size_t read_cnt = fread(&size, sizeof(size_t), 1, stream);
|
|
|
+ if (read_cnt != 1) {
|
|
|
+ throw std::runtime_error("Cannot read from file");
|
|
|
+ }
|
|
|
+ value.resize(size);
|
|
|
+ read_cnt = fread(&value[0], sizeof(T), size, stream);
|
|
|
+ if (read_cnt != size) {
|
|
|
+ throw std::runtime_error("Cannot read from file");
|
|
|
+ }
|
|
|
+}
|
|
|
+/** @} */
|
|
|
+
|
|
|
+/** @addtogroup metric_grp Metric (distance) classes
|
|
|
+ * @{ */
|
|
|
+
|
|
|
+struct Metric {};
|
|
|
+
|
|
|
+/** Manhattan distance functor (generic version, optimized for
|
|
|
+ * high-dimensionality data sets). Corresponding distance traits:
|
|
|
+ * nanoflann::metric_L1 \tparam T Type of the elements (e.g. double, float,
|
|
|
+ * uint8_t) \tparam _DistanceType Type of distance variables (must be signed)
|
|
|
+ * (e.g. float, double, int64_t)
|
|
|
+ */
|
|
|
+template <class T, class DataSource, typename _DistanceType = T>
|
|
|
+struct L1_Adaptor {
|
|
|
+ typedef T ElementType;
|
|
|
+ typedef _DistanceType DistanceType;
|
|
|
+
|
|
|
+ const DataSource &data_source;
|
|
|
+
|
|
|
+ L1_Adaptor(const DataSource &_data_source) : data_source(_data_source) {}
|
|
|
+
|
|
|
+ inline DistanceType evalMetric(const T *a, const size_t b_idx, size_t size,
|
|
|
+ DistanceType worst_dist = -1) const {
|
|
|
+ DistanceType result = DistanceType();
|
|
|
+ const T *last = a + size;
|
|
|
+ const T *lastgroup = last - 3;
|
|
|
+ size_t d = 0;
|
|
|
+
|
|
|
+ /* Process 4 items with each loop for efficiency. */
|
|
|
+ while (a < lastgroup) {
|
|
|
+ const DistanceType diff0 =
|
|
|
+ std::abs(a[0] - data_source.kdtree_get_pt(b_idx, d++));
|
|
|
+ const DistanceType diff1 =
|
|
|
+ std::abs(a[1] - data_source.kdtree_get_pt(b_idx, d++));
|
|
|
+ const DistanceType diff2 =
|
|
|
+ std::abs(a[2] - data_source.kdtree_get_pt(b_idx, d++));
|
|
|
+ const DistanceType diff3 =
|
|
|
+ std::abs(a[3] - data_source.kdtree_get_pt(b_idx, d++));
|
|
|
+ result += diff0 + diff1 + diff2 + diff3;
|
|
|
+ a += 4;
|
|
|
+ if ((worst_dist > 0) && (result > worst_dist)) {
|
|
|
+ return result;
|
|
|
+ }
|
|
|
+ }
|
|
|
+ /* Process last 0-3 components. Not needed for standard vector lengths. */
|
|
|
+ while (a < last) {
|
|
|
+ result += std::abs(*a++ - data_source.kdtree_get_pt(b_idx, d++));
|
|
|
+ }
|
|
|
+ return result;
|
|
|
+ }
|
|
|
+
|
|
|
+ template <typename U, typename V>
|
|
|
+ inline DistanceType accum_dist(const U a, const V b, const size_t) const {
|
|
|
+ return std::abs(a - b);
|
|
|
+ }
|
|
|
+};
|
|
|
+
|
|
|
+/** Squared Euclidean distance functor (generic version, optimized for
|
|
|
+ * high-dimensionality data sets). Corresponding distance traits:
|
|
|
+ * nanoflann::metric_L2 \tparam T Type of the elements (e.g. double, float,
|
|
|
+ * uint8_t) \tparam _DistanceType Type of distance variables (must be signed)
|
|
|
+ * (e.g. float, double, int64_t)
|
|
|
+ */
|
|
|
+template <class T, class DataSource, typename _DistanceType = T>
|
|
|
+struct L2_Adaptor {
|
|
|
+ typedef T ElementType;
|
|
|
+ typedef _DistanceType DistanceType;
|
|
|
+
|
|
|
+ const DataSource &data_source;
|
|
|
+
|
|
|
+ L2_Adaptor(const DataSource &_data_source) : data_source(_data_source) {}
|
|
|
+
|
|
|
+ inline DistanceType evalMetric(const T *a, const size_t b_idx, size_t size,
|
|
|
+ DistanceType worst_dist = -1) const {
|
|
|
+ DistanceType result = DistanceType();
|
|
|
+ const T *last = a + size;
|
|
|
+ const T *lastgroup = last - 3;
|
|
|
+ size_t d = 0;
|
|
|
+
|
|
|
+ /* Process 4 items with each loop for efficiency. */
|
|
|
+ while (a < lastgroup) {
|
|
|
+ const DistanceType diff0 = a[0] - data_source.kdtree_get_pt(b_idx, d++);
|
|
|
+ const DistanceType diff1 = a[1] - data_source.kdtree_get_pt(b_idx, d++);
|
|
|
+ const DistanceType diff2 = a[2] - data_source.kdtree_get_pt(b_idx, d++);
|
|
|
+ const DistanceType diff3 = a[3] - data_source.kdtree_get_pt(b_idx, d++);
|
|
|
+ result += diff0 * diff0 + diff1 * diff1 + diff2 * diff2 + diff3 * diff3;
|
|
|
+ a += 4;
|
|
|
+ if ((worst_dist > 0) && (result > worst_dist)) {
|
|
|
+ return result;
|
|
|
+ }
|
|
|
+ }
|
|
|
+ /* Process last 0-3 components. Not needed for standard vector lengths. */
|
|
|
+ while (a < last) {
|
|
|
+ const DistanceType diff0 = *a++ - data_source.kdtree_get_pt(b_idx, d++);
|
|
|
+ result += diff0 * diff0;
|
|
|
+ }
|
|
|
+ return result;
|
|
|
+ }
|
|
|
+
|
|
|
+ template <typename U, typename V>
|
|
|
+ inline DistanceType accum_dist(const U a, const V b, const size_t) const {
|
|
|
+ return (a - b) * (a - b);
|
|
|
+ }
|
|
|
+};
|
|
|
+
|
|
|
+/** Squared Euclidean (L2) distance functor (suitable for low-dimensionality
|
|
|
+ * datasets, like 2D or 3D point clouds) Corresponding distance traits:
|
|
|
+ * nanoflann::metric_L2_Simple \tparam T Type of the elements (e.g. double,
|
|
|
+ * float, uint8_t) \tparam _DistanceType Type of distance variables (must be
|
|
|
+ * signed) (e.g. float, double, int64_t)
|
|
|
+ */
|
|
|
+template <class T, class DataSource, typename _DistanceType = T>
|
|
|
+struct L2_Simple_Adaptor {
|
|
|
+ typedef T ElementType;
|
|
|
+ typedef _DistanceType DistanceType;
|
|
|
+
|
|
|
+ const DataSource &data_source;
|
|
|
+
|
|
|
+ L2_Simple_Adaptor(const DataSource &_data_source)
|
|
|
+ : data_source(_data_source) {}
|
|
|
+
|
|
|
+ inline DistanceType evalMetric(const T *a, const size_t b_idx,
|
|
|
+ size_t size) const {
|
|
|
+ DistanceType result = DistanceType();
|
|
|
+ for (size_t i = 0; i < size; ++i) {
|
|
|
+ const DistanceType diff = a[i] - data_source.kdtree_get_pt(b_idx, i);
|
|
|
+ result += diff * diff;
|
|
|
+ }
|
|
|
+ return result;
|
|
|
+ }
|
|
|
+
|
|
|
+ template <typename U, typename V>
|
|
|
+ inline DistanceType accum_dist(const U a, const V b, const size_t) const {
|
|
|
+ return (a - b) * (a - b);
|
|
|
+ }
|
|
|
+};
|
|
|
+
|
|
|
+/** SO2 distance functor
|
|
|
+ * Corresponding distance traits: nanoflann::metric_SO2
|
|
|
+ * \tparam T Type of the elements (e.g. double, float)
|
|
|
+ * \tparam _DistanceType Type of distance variables (must be signed) (e.g.
|
|
|
+ * float, double) orientation is constrained to be in [-pi, pi]
|
|
|
+ */
|
|
|
+template <class T, class DataSource, typename _DistanceType = T>
|
|
|
+struct SO2_Adaptor {
|
|
|
+ typedef T ElementType;
|
|
|
+ typedef _DistanceType DistanceType;
|
|
|
+
|
|
|
+ const DataSource &data_source;
|
|
|
+
|
|
|
+ SO2_Adaptor(const DataSource &_data_source) : data_source(_data_source) {}
|
|
|
+
|
|
|
+ inline DistanceType evalMetric(const T *a, const size_t b_idx,
|
|
|
+ size_t size) const {
|
|
|
+ return accum_dist(a[size - 1], data_source.kdtree_get_pt(b_idx, size - 1),
|
|
|
+ size - 1);
|
|
|
+ }
|
|
|
+
|
|
|
+ /** Note: this assumes that input angles are already in the range [-pi,pi] */
|
|
|
+ template <typename U, typename V>
|
|
|
+ inline DistanceType accum_dist(const U a, const V b, const size_t) const {
|
|
|
+ DistanceType result = DistanceType();
|
|
|
+ DistanceType PI = pi_const<DistanceType>();
|
|
|
+ result = b - a;
|
|
|
+ if (result > PI)
|
|
|
+ result -= 2 * PI;
|
|
|
+ else if (result < -PI)
|
|
|
+ result += 2 * PI;
|
|
|
+ return result;
|
|
|
+ }
|
|
|
+};
|
|
|
+
|
|
|
+/** SO3 distance functor (Uses L2_Simple)
|
|
|
+ * Corresponding distance traits: nanoflann::metric_SO3
|
|
|
+ * \tparam T Type of the elements (e.g. double, float)
|
|
|
+ * \tparam _DistanceType Type of distance variables (must be signed) (e.g.
|
|
|
+ * float, double)
|
|
|
+ */
|
|
|
+template <class T, class DataSource, typename _DistanceType = T>
|
|
|
+struct SO3_Adaptor {
|
|
|
+ typedef T ElementType;
|
|
|
+ typedef _DistanceType DistanceType;
|
|
|
+
|
|
|
+ L2_Simple_Adaptor<T, DataSource> distance_L2_Simple;
|
|
|
+
|
|
|
+ SO3_Adaptor(const DataSource &_data_source)
|
|
|
+ : distance_L2_Simple(_data_source) {}
|
|
|
+
|
|
|
+ inline DistanceType evalMetric(const T *a, const size_t b_idx,
|
|
|
+ size_t size) const {
|
|
|
+ return distance_L2_Simple.evalMetric(a, b_idx, size);
|
|
|
+ }
|
|
|
+
|
|
|
+ template <typename U, typename V>
|
|
|
+ inline DistanceType accum_dist(const U a, const V b, const size_t idx) const {
|
|
|
+ return distance_L2_Simple.accum_dist(a, b, idx);
|
|
|
+ }
|
|
|
+};
|
|
|
+
|
|
|
+/** Metaprogramming helper traits class for the L1 (Manhattan) metric */
|
|
|
+struct metric_L1 : public Metric {
|
|
|
+ template <class T, class DataSource> struct traits {
|
|
|
+ typedef L1_Adaptor<T, DataSource> distance_t;
|
|
|
+ };
|
|
|
+};
|
|
|
+/** Metaprogramming helper traits class for the L2 (Euclidean) metric */
|
|
|
+struct metric_L2 : public Metric {
|
|
|
+ template <class T, class DataSource> struct traits {
|
|
|
+ typedef L2_Adaptor<T, DataSource> distance_t;
|
|
|
+ };
|
|
|
+};
|
|
|
+/** Metaprogramming helper traits class for the L2_simple (Euclidean) metric */
|
|
|
+struct metric_L2_Simple : public Metric {
|
|
|
+ template <class T, class DataSource> struct traits {
|
|
|
+ typedef L2_Simple_Adaptor<T, DataSource> distance_t;
|
|
|
+ };
|
|
|
+};
|
|
|
+/** Metaprogramming helper traits class for the SO3_InnerProdQuat metric */
|
|
|
+struct metric_SO2 : public Metric {
|
|
|
+ template <class T, class DataSource> struct traits {
|
|
|
+ typedef SO2_Adaptor<T, DataSource> distance_t;
|
|
|
+ };
|
|
|
+};
|
|
|
+/** Metaprogramming helper traits class for the SO3_InnerProdQuat metric */
|
|
|
+struct metric_SO3 : public Metric {
|
|
|
+ template <class T, class DataSource> struct traits {
|
|
|
+ typedef SO3_Adaptor<T, DataSource> distance_t;
|
|
|
+ };
|
|
|
+};
|
|
|
+
|
|
|
+/** @} */
|
|
|
+
|
|
|
+/** @addtogroup param_grp Parameter structs
|
|
|
+ * @{ */
|
|
|
+
|
|
|
+/** Parameters (see README.md) */
|
|
|
+struct KDTreeSingleIndexAdaptorParams {
|
|
|
+ KDTreeSingleIndexAdaptorParams(size_t _leaf_max_size = 10)
|
|
|
+ : leaf_max_size(_leaf_max_size) {}
|
|
|
+
|
|
|
+ size_t leaf_max_size;
|
|
|
+};
|
|
|
+
|
|
|
+/** Search options for KDTreeSingleIndexAdaptor::findNeighbors() */
|
|
|
+struct SearchParams {
|
|
|
+ /** Note: The first argument (checks_IGNORED_) is ignored, but kept for
|
|
|
+ * compatibility with the FLANN interface */
|
|
|
+ SearchParams(int checks_IGNORED_ = 32, float eps_ = 0, bool sorted_ = true)
|
|
|
+ : checks(checks_IGNORED_), eps(eps_), sorted(sorted_) {}
|
|
|
+
|
|
|
+ int checks; //!< Ignored parameter (Kept for compatibility with the FLANN
|
|
|
+ //!< interface).
|
|
|
+ float eps; //!< search for eps-approximate neighbours (default: 0)
|
|
|
+ bool sorted; //!< only for radius search, require neighbours sorted by
|
|
|
+ //!< distance (default: true)
|
|
|
+};
|
|
|
+/** @} */
|
|
|
+
|
|
|
+/** @addtogroup memalloc_grp Memory allocation
|
|
|
+ * @{ */
|
|
|
+
|
|
|
+/**
|
|
|
+ * Allocates (using C's malloc) a generic type T.
|
|
|
+ *
|
|
|
+ * Params:
|
|
|
+ * count = number of instances to allocate.
|
|
|
+ * Returns: pointer (of type T*) to memory buffer
|
|
|
+ */
|
|
|
+template <typename T> inline T *allocate(size_t count = 1) {
|
|
|
+ T *mem = static_cast<T *>(::malloc(sizeof(T) * count));
|
|
|
+ return mem;
|
|
|
+}
|
|
|
+
|
|
|
+/**
|
|
|
+ * Pooled storage allocator
|
|
|
+ *
|
|
|
+ * The following routines allow for the efficient allocation of storage in
|
|
|
+ * small chunks from a specified pool. Rather than allowing each structure
|
|
|
+ * to be freed individually, an entire pool of storage is freed at once.
|
|
|
+ * This method has two advantages over just using malloc() and free(). First,
|
|
|
+ * it is far more efficient for allocating small objects, as there is
|
|
|
+ * no overhead for remembering all the information needed to free each
|
|
|
+ * object or consolidating fragmented memory. Second, the decision about
|
|
|
+ * how long to keep an object is made at the time of allocation, and there
|
|
|
+ * is no need to track down all the objects to free them.
|
|
|
+ *
|
|
|
+ */
|
|
|
+
|
|
|
+const size_t WORDSIZE = 16;
|
|
|
+const size_t BLOCKSIZE = 8192;
|
|
|
+
|
|
|
+class PooledAllocator {
|
|
|
+ /* We maintain memory alignment to word boundaries by requiring that all
|
|
|
+ allocations be in multiples of the machine wordsize. */
|
|
|
+ /* Size of machine word in bytes. Must be power of 2. */
|
|
|
+ /* Minimum number of bytes requested at a time from the system. Must be
|
|
|
+ * multiple of WORDSIZE. */
|
|
|
+
|
|
|
+ size_t remaining; /* Number of bytes left in current block of storage. */
|
|
|
+ void *base; /* Pointer to base of current block of storage. */
|
|
|
+ void *loc; /* Current location in block to next allocate memory. */
|
|
|
+
|
|
|
+ void internal_init() {
|
|
|
+ remaining = 0;
|
|
|
+ base = NULL;
|
|
|
+ usedMemory = 0;
|
|
|
+ wastedMemory = 0;
|
|
|
+ }
|
|
|
+
|
|
|
+public:
|
|
|
+ size_t usedMemory;
|
|
|
+ size_t wastedMemory;
|
|
|
+
|
|
|
+ /**
|
|
|
+ Default constructor. Initializes a new pool.
|
|
|
+ */
|
|
|
+ PooledAllocator() { internal_init(); }
|
|
|
+
|
|
|
+ /**
|
|
|
+ * Destructor. Frees all the memory allocated in this pool.
|
|
|
+ */
|
|
|
+ ~PooledAllocator() { free_all(); }
|
|
|
+
|
|
|
+ /** Frees all allocated memory chunks */
|
|
|
+ void free_all() {
|
|
|
+ while (base != NULL) {
|
|
|
+ void *prev =
|
|
|
+ *(static_cast<void **>(base)); /* Get pointer to prev block. */
|
|
|
+ ::free(base);
|
|
|
+ base = prev;
|
|
|
+ }
|
|
|
+ internal_init();
|
|
|
+ }
|
|
|
+
|
|
|
+ /**
|
|
|
+ * Returns a pointer to a piece of new memory of the given size in bytes
|
|
|
+ * allocated from the pool.
|
|
|
+ */
|
|
|
+ void *malloc(const size_t req_size) {
|
|
|
+ /* Round size up to a multiple of wordsize. The following expression
|
|
|
+ only works for WORDSIZE that is a power of 2, by masking last bits of
|
|
|
+ incremented size to zero.
|
|
|
+ */
|
|
|
+ const size_t size = (req_size + (WORDSIZE - 1)) & ~(WORDSIZE - 1);
|
|
|
+
|
|
|
+ /* Check whether a new block must be allocated. Note that the first word
|
|
|
+ of a block is reserved for a pointer to the previous block.
|
|
|
+ */
|
|
|
+ if (size > remaining) {
|
|
|
+
|
|
|
+ wastedMemory += remaining;
|
|
|
+
|
|
|
+ /* Allocate new storage. */
|
|
|
+ const size_t blocksize =
|
|
|
+ (size + sizeof(void *) + (WORDSIZE - 1) > BLOCKSIZE)
|
|
|
+ ? size + sizeof(void *) + (WORDSIZE - 1)
|
|
|
+ : BLOCKSIZE;
|
|
|
+
|
|
|
+ // use the standard C malloc to allocate memory
|
|
|
+ void *m = ::malloc(blocksize);
|
|
|
+ if (!m) {
|
|
|
+ fprintf(stderr, "Failed to allocate memory.\n");
|
|
|
+ throw std::bad_alloc();
|
|
|
+ }
|
|
|
+
|
|
|
+ /* Fill first word of new block with pointer to previous block. */
|
|
|
+ static_cast<void **>(m)[0] = base;
|
|
|
+ base = m;
|
|
|
+
|
|
|
+ size_t shift = 0;
|
|
|
+ // int size_t = (WORDSIZE - ( (((size_t)m) + sizeof(void*)) &
|
|
|
+ // (WORDSIZE-1))) & (WORDSIZE-1);
|
|
|
+
|
|
|
+ remaining = blocksize - sizeof(void *) - shift;
|
|
|
+ loc = (static_cast<char *>(m) + sizeof(void *) + shift);
|
|
|
+ }
|
|
|
+ void *rloc = loc;
|
|
|
+ loc = static_cast<char *>(loc) + size;
|
|
|
+ remaining -= size;
|
|
|
+
|
|
|
+ usedMemory += size;
|
|
|
+
|
|
|
+ return rloc;
|
|
|
+ }
|
|
|
+
|
|
|
+ /**
|
|
|
+ * Allocates (using this pool) a generic type T.
|
|
|
+ *
|
|
|
+ * Params:
|
|
|
+ * count = number of instances to allocate.
|
|
|
+ * Returns: pointer (of type T*) to memory buffer
|
|
|
+ */
|
|
|
+ template <typename T> T *allocate(const size_t count = 1) {
|
|
|
+ T *mem = static_cast<T *>(this->malloc(sizeof(T) * count));
|
|
|
+ return mem;
|
|
|
+ }
|
|
|
+};
|
|
|
+/** @} */
|
|
|
+
|
|
|
+/** @addtogroup nanoflann_metaprog_grp Auxiliary metaprogramming stuff
|
|
|
+ * @{ */
|
|
|
+
|
|
|
+/** Used to declare fixed-size arrays when DIM>0, dynamically-allocated vectors
|
|
|
+ * when DIM=-1. Fixed size version for a generic DIM:
|
|
|
+ */
|
|
|
+template <int DIM, typename T> struct array_or_vector_selector {
|
|
|
+ typedef std::array<T, DIM> container_t;
|
|
|
+};
|
|
|
+/** Dynamic size version */
|
|
|
+template <typename T> struct array_or_vector_selector<-1, T> {
|
|
|
+ typedef std::vector<T> container_t;
|
|
|
+};
|
|
|
+
|
|
|
+/** @} */
|
|
|
+
|
|
|
+/** kd-tree base-class
|
|
|
+ *
|
|
|
+ * Contains the member functions common to the classes KDTreeSingleIndexAdaptor
|
|
|
+ * and KDTreeSingleIndexDynamicAdaptor_.
|
|
|
+ *
|
|
|
+ * \tparam Derived The name of the class which inherits this class.
|
|
|
+ * \tparam DatasetAdaptor The user-provided adaptor (see comments above).
|
|
|
+ * \tparam Distance The distance metric to use, these are all classes derived
|
|
|
+ * from nanoflann::Metric \tparam DIM Dimensionality of data points (e.g. 3 for
|
|
|
+ * 3D points) \tparam IndexType Will be typically size_t or int
|
|
|
+ */
|
|
|
+
|
|
|
+template <class Derived, typename Distance, class DatasetAdaptor, int DIM = -1,
|
|
|
+ typename IndexType = size_t>
|
|
|
+class KDTreeBaseClass {
|
|
|
+
|
|
|
+public:
|
|
|
+ /** Frees the previously-built index. Automatically called within
|
|
|
+ * buildIndex(). */
|
|
|
+ void freeIndex(Derived &obj) {
|
|
|
+ obj.pool.free_all();
|
|
|
+ obj.root_node = NULL;
|
|
|
+ obj.m_size_at_index_build = 0;
|
|
|
+ }
|
|
|
+
|
|
|
+ typedef typename Distance::ElementType ElementType;
|
|
|
+ typedef typename Distance::DistanceType DistanceType;
|
|
|
+
|
|
|
+ /*--------------------- Internal Data Structures --------------------------*/
|
|
|
+ struct Node {
|
|
|
+ /** Union used because a node can be either a LEAF node or a non-leaf node,
|
|
|
+ * so both data fields are never used simultaneously */
|
|
|
+ union {
|
|
|
+ struct leaf {
|
|
|
+ IndexType left, right; //!< Indices of points in leaf node
|
|
|
+ } lr;
|
|
|
+ struct nonleaf {
|
|
|
+ int divfeat; //!< Dimension used for subdivision.
|
|
|
+ DistanceType divlow, divhigh; //!< The values used for subdivision.
|
|
|
+ } sub;
|
|
|
+ } node_type;
|
|
|
+ Node *child1, *child2; //!< Child nodes (both=NULL mean its a leaf node)
|
|
|
+ };
|
|
|
+
|
|
|
+ typedef Node *NodePtr;
|
|
|
+
|
|
|
+ struct Interval {
|
|
|
+ ElementType low, high;
|
|
|
+ };
|
|
|
+
|
|
|
+ /**
|
|
|
+ * Array of indices to vectors in the dataset.
|
|
|
+ */
|
|
|
+ std::vector<IndexType> vind;
|
|
|
+
|
|
|
+ NodePtr root_node;
|
|
|
+
|
|
|
+ size_t m_leaf_max_size;
|
|
|
+
|
|
|
+ size_t m_size; //!< Number of current points in the dataset
|
|
|
+ size_t m_size_at_index_build; //!< Number of points in the dataset when the
|
|
|
+ //!< index was built
|
|
|
+ int dim; //!< Dimensionality of each data point
|
|
|
+
|
|
|
+ /** Define "BoundingBox" as a fixed-size or variable-size container depending
|
|
|
+ * on "DIM" */
|
|
|
+ typedef
|
|
|
+ typename array_or_vector_selector<DIM, Interval>::container_t BoundingBox;
|
|
|
+
|
|
|
+ /** Define "distance_vector_t" as a fixed-size or variable-size container
|
|
|
+ * depending on "DIM" */
|
|
|
+ typedef typename array_or_vector_selector<DIM, DistanceType>::container_t
|
|
|
+ distance_vector_t;
|
|
|
+
|
|
|
+ /** The KD-tree used to find neighbours */
|
|
|
+
|
|
|
+ BoundingBox root_bbox;
|
|
|
+
|
|
|
+ /**
|
|
|
+ * Pooled memory allocator.
|
|
|
+ *
|
|
|
+ * Using a pooled memory allocator is more efficient
|
|
|
+ * than allocating memory directly when there is a large
|
|
|
+ * number small of memory allocations.
|
|
|
+ */
|
|
|
+ PooledAllocator pool;
|
|
|
+
|
|
|
+ /** Returns number of points in dataset */
|
|
|
+ size_t size(const Derived &obj) const { return obj.m_size; }
|
|
|
+
|
|
|
+ /** Returns the length of each point in the dataset */
|
|
|
+ size_t veclen(const Derived &obj) {
|
|
|
+ return static_cast<size_t>(DIM > 0 ? DIM : obj.dim);
|
|
|
+ }
|
|
|
+
|
|
|
+ /// Helper accessor to the dataset points:
|
|
|
+ inline ElementType dataset_get(const Derived &obj, size_t idx,
|
|
|
+ int component) const {
|
|
|
+ return obj.dataset.kdtree_get_pt(idx, component);
|
|
|
+ }
|
|
|
+
|
|
|
+ /**
|
|
|
+ * Computes the inde memory usage
|
|
|
+ * Returns: memory used by the index
|
|
|
+ */
|
|
|
+ size_t usedMemory(Derived &obj) {
|
|
|
+ return obj.pool.usedMemory + obj.pool.wastedMemory +
|
|
|
+ obj.dataset.kdtree_get_point_count() *
|
|
|
+ sizeof(IndexType); // pool memory and vind array memory
|
|
|
+ }
|
|
|
+
|
|
|
+ void computeMinMax(const Derived &obj, IndexType *ind, IndexType count,
|
|
|
+ int element, ElementType &min_elem,
|
|
|
+ ElementType &max_elem) {
|
|
|
+ min_elem = dataset_get(obj, ind[0], element);
|
|
|
+ max_elem = dataset_get(obj, ind[0], element);
|
|
|
+ for (IndexType i = 1; i < count; ++i) {
|
|
|
+ ElementType val = dataset_get(obj, ind[i], element);
|
|
|
+ if (val < min_elem)
|
|
|
+ min_elem = val;
|
|
|
+ if (val > max_elem)
|
|
|
+ max_elem = val;
|
|
|
+ }
|
|
|
+ }
|
|
|
+
|
|
|
+ /**
|
|
|
+ * Create a tree node that subdivides the list of vecs from vind[first]
|
|
|
+ * to vind[last]. The routine is called recursively on each sublist.
|
|
|
+ *
|
|
|
+ * @param left index of the first vector
|
|
|
+ * @param right index of the last vector
|
|
|
+ */
|
|
|
+ NodePtr divideTree(Derived &obj, const IndexType left, const IndexType right,
|
|
|
+ BoundingBox &bbox) {
|
|
|
+ NodePtr node = obj.pool.template allocate<Node>(); // allocate memory
|
|
|
+
|
|
|
+ /* If too few exemplars remain, then make this a leaf node. */
|
|
|
+ if ((right - left) <= static_cast<IndexType>(obj.m_leaf_max_size)) {
|
|
|
+ node->child1 = node->child2 = NULL; /* Mark as leaf node. */
|
|
|
+ node->node_type.lr.left = left;
|
|
|
+ node->node_type.lr.right = right;
|
|
|
+
|
|
|
+ // compute bounding-box of leaf points
|
|
|
+ for (int i = 0; i < (DIM > 0 ? DIM : obj.dim); ++i) {
|
|
|
+ bbox[i].low = dataset_get(obj, obj.vind[left], i);
|
|
|
+ bbox[i].high = dataset_get(obj, obj.vind[left], i);
|
|
|
+ }
|
|
|
+ for (IndexType k = left + 1; k < right; ++k) {
|
|
|
+ for (int i = 0; i < (DIM > 0 ? DIM : obj.dim); ++i) {
|
|
|
+ if (bbox[i].low > dataset_get(obj, obj.vind[k], i))
|
|
|
+ bbox[i].low = dataset_get(obj, obj.vind[k], i);
|
|
|
+ if (bbox[i].high < dataset_get(obj, obj.vind[k], i))
|
|
|
+ bbox[i].high = dataset_get(obj, obj.vind[k], i);
|
|
|
+ }
|
|
|
+ }
|
|
|
+ } else {
|
|
|
+ IndexType idx;
|
|
|
+ int cutfeat;
|
|
|
+ DistanceType cutval;
|
|
|
+ middleSplit_(obj, &obj.vind[0] + left, right - left, idx, cutfeat, cutval,
|
|
|
+ bbox);
|
|
|
+
|
|
|
+ node->node_type.sub.divfeat = cutfeat;
|
|
|
+
|
|
|
+ BoundingBox left_bbox(bbox);
|
|
|
+ left_bbox[cutfeat].high = cutval;
|
|
|
+ node->child1 = divideTree(obj, left, left + idx, left_bbox);
|
|
|
+
|
|
|
+ BoundingBox right_bbox(bbox);
|
|
|
+ right_bbox[cutfeat].low = cutval;
|
|
|
+ node->child2 = divideTree(obj, left + idx, right, right_bbox);
|
|
|
+
|
|
|
+ node->node_type.sub.divlow = left_bbox[cutfeat].high;
|
|
|
+ node->node_type.sub.divhigh = right_bbox[cutfeat].low;
|
|
|
+
|
|
|
+ for (int i = 0; i < (DIM > 0 ? DIM : obj.dim); ++i) {
|
|
|
+ bbox[i].low = std::min(left_bbox[i].low, right_bbox[i].low);
|
|
|
+ bbox[i].high = std::max(left_bbox[i].high, right_bbox[i].high);
|
|
|
+ }
|
|
|
+ }
|
|
|
+
|
|
|
+ return node;
|
|
|
+ }
|
|
|
+
|
|
|
+ void middleSplit_(Derived &obj, IndexType *ind, IndexType count,
|
|
|
+ IndexType &index, int &cutfeat, DistanceType &cutval,
|
|
|
+ const BoundingBox &bbox) {
|
|
|
+ const DistanceType EPS = static_cast<DistanceType>(0.00001);
|
|
|
+ ElementType max_span = bbox[0].high - bbox[0].low;
|
|
|
+ for (int i = 1; i < (DIM > 0 ? DIM : obj.dim); ++i) {
|
|
|
+ ElementType span = bbox[i].high - bbox[i].low;
|
|
|
+ if (span > max_span) {
|
|
|
+ max_span = span;
|
|
|
+ }
|
|
|
+ }
|
|
|
+ ElementType max_spread = -1;
|
|
|
+ cutfeat = 0;
|
|
|
+ for (int i = 0; i < (DIM > 0 ? DIM : obj.dim); ++i) {
|
|
|
+ ElementType span = bbox[i].high - bbox[i].low;
|
|
|
+ if (span > (1 - EPS) * max_span) {
|
|
|
+ ElementType min_elem, max_elem;
|
|
|
+ computeMinMax(obj, ind, count, i, min_elem, max_elem);
|
|
|
+ ElementType spread = max_elem - min_elem;
|
|
|
+ if (spread > max_spread) {
|
|
|
+ cutfeat = i;
|
|
|
+ max_spread = spread;
|
|
|
+ }
|
|
|
+ }
|
|
|
+ }
|
|
|
+ // split in the middle
|
|
|
+ DistanceType split_val = (bbox[cutfeat].low + bbox[cutfeat].high) / 2;
|
|
|
+ ElementType min_elem, max_elem;
|
|
|
+ computeMinMax(obj, ind, count, cutfeat, min_elem, max_elem);
|
|
|
+
|
|
|
+ if (split_val < min_elem)
|
|
|
+ cutval = min_elem;
|
|
|
+ else if (split_val > max_elem)
|
|
|
+ cutval = max_elem;
|
|
|
+ else
|
|
|
+ cutval = split_val;
|
|
|
+
|
|
|
+ IndexType lim1, lim2;
|
|
|
+ planeSplit(obj, ind, count, cutfeat, cutval, lim1, lim2);
|
|
|
+
|
|
|
+ if (lim1 > count / 2)
|
|
|
+ index = lim1;
|
|
|
+ else if (lim2 < count / 2)
|
|
|
+ index = lim2;
|
|
|
+ else
|
|
|
+ index = count / 2;
|
|
|
+ }
|
|
|
+
|
|
|
+ /**
|
|
|
+ * Subdivide the list of points by a plane perpendicular on axe corresponding
|
|
|
+ * to the 'cutfeat' dimension at 'cutval' position.
|
|
|
+ *
|
|
|
+ * On return:
|
|
|
+ * dataset[ind[0..lim1-1]][cutfeat]<cutval
|
|
|
+ * dataset[ind[lim1..lim2-1]][cutfeat]==cutval
|
|
|
+ * dataset[ind[lim2..count]][cutfeat]>cutval
|
|
|
+ */
|
|
|
+ void planeSplit(Derived &obj, IndexType *ind, const IndexType count,
|
|
|
+ int cutfeat, DistanceType &cutval, IndexType &lim1,
|
|
|
+ IndexType &lim2) {
|
|
|
+ /* Move vector indices for left subtree to front of list. */
|
|
|
+ IndexType left = 0;
|
|
|
+ IndexType right = count - 1;
|
|
|
+ for (;;) {
|
|
|
+ while (left <= right && dataset_get(obj, ind[left], cutfeat) < cutval)
|
|
|
+ ++left;
|
|
|
+ while (right && left <= right &&
|
|
|
+ dataset_get(obj, ind[right], cutfeat) >= cutval)
|
|
|
+ --right;
|
|
|
+ if (left > right || !right)
|
|
|
+ break; // "!right" was added to support unsigned Index types
|
|
|
+ std::swap(ind[left], ind[right]);
|
|
|
+ ++left;
|
|
|
+ --right;
|
|
|
+ }
|
|
|
+ /* If either list is empty, it means that all remaining features
|
|
|
+ * are identical. Split in the middle to maintain a balanced tree.
|
|
|
+ */
|
|
|
+ lim1 = left;
|
|
|
+ right = count - 1;
|
|
|
+ for (;;) {
|
|
|
+ while (left <= right && dataset_get(obj, ind[left], cutfeat) <= cutval)
|
|
|
+ ++left;
|
|
|
+ while (right && left <= right &&
|
|
|
+ dataset_get(obj, ind[right], cutfeat) > cutval)
|
|
|
+ --right;
|
|
|
+ if (left > right || !right)
|
|
|
+ break; // "!right" was added to support unsigned Index types
|
|
|
+ std::swap(ind[left], ind[right]);
|
|
|
+ ++left;
|
|
|
+ --right;
|
|
|
+ }
|
|
|
+ lim2 = left;
|
|
|
+ }
|
|
|
+
|
|
|
+ DistanceType computeInitialDistances(const Derived &obj,
|
|
|
+ const ElementType *vec,
|
|
|
+ distance_vector_t &dists) const {
|
|
|
+ assert(vec);
|
|
|
+ DistanceType distsq = DistanceType();
|
|
|
+
|
|
|
+ for (int i = 0; i < (DIM > 0 ? DIM : obj.dim); ++i) {
|
|
|
+ if (vec[i] < obj.root_bbox[i].low) {
|
|
|
+ dists[i] = obj.distance.accum_dist(vec[i], obj.root_bbox[i].low, i);
|
|
|
+ distsq += dists[i];
|
|
|
+ }
|
|
|
+ if (vec[i] > obj.root_bbox[i].high) {
|
|
|
+ dists[i] = obj.distance.accum_dist(vec[i], obj.root_bbox[i].high, i);
|
|
|
+ distsq += dists[i];
|
|
|
+ }
|
|
|
+ }
|
|
|
+ return distsq;
|
|
|
+ }
|
|
|
+
|
|
|
+ void save_tree(Derived &obj, FILE *stream, NodePtr tree) {
|
|
|
+ save_value(stream, *tree);
|
|
|
+ if (tree->child1 != NULL) {
|
|
|
+ save_tree(obj, stream, tree->child1);
|
|
|
+ }
|
|
|
+ if (tree->child2 != NULL) {
|
|
|
+ save_tree(obj, stream, tree->child2);
|
|
|
+ }
|
|
|
+ }
|
|
|
+
|
|
|
+ void load_tree(Derived &obj, FILE *stream, NodePtr &tree) {
|
|
|
+ tree = obj.pool.template allocate<Node>();
|
|
|
+ load_value(stream, *tree);
|
|
|
+ if (tree->child1 != NULL) {
|
|
|
+ load_tree(obj, stream, tree->child1);
|
|
|
+ }
|
|
|
+ if (tree->child2 != NULL) {
|
|
|
+ load_tree(obj, stream, tree->child2);
|
|
|
+ }
|
|
|
+ }
|
|
|
+
|
|
|
+ /** Stores the index in a binary file.
|
|
|
+ * IMPORTANT NOTE: The set of data points is NOT stored in the file, so when
|
|
|
+ * loading the index object it must be constructed associated to the same
|
|
|
+ * source of data points used while building it. See the example:
|
|
|
+ * examples/saveload_example.cpp \sa loadIndex */
|
|
|
+ void saveIndex_(Derived &obj, FILE *stream) {
|
|
|
+ save_value(stream, obj.m_size);
|
|
|
+ save_value(stream, obj.dim);
|
|
|
+ save_value(stream, obj.root_bbox);
|
|
|
+ save_value(stream, obj.m_leaf_max_size);
|
|
|
+ save_value(stream, obj.vind);
|
|
|
+ save_tree(obj, stream, obj.root_node);
|
|
|
+ }
|
|
|
+
|
|
|
+ /** Loads a previous index from a binary file.
|
|
|
+ * IMPORTANT NOTE: The set of data points is NOT stored in the file, so the
|
|
|
+ * index object must be constructed associated to the same source of data
|
|
|
+ * points used while building the index. See the example:
|
|
|
+ * examples/saveload_example.cpp \sa loadIndex */
|
|
|
+ void loadIndex_(Derived &obj, FILE *stream) {
|
|
|
+ load_value(stream, obj.m_size);
|
|
|
+ load_value(stream, obj.dim);
|
|
|
+ load_value(stream, obj.root_bbox);
|
|
|
+ load_value(stream, obj.m_leaf_max_size);
|
|
|
+ load_value(stream, obj.vind);
|
|
|
+ load_tree(obj, stream, obj.root_node);
|
|
|
+ }
|
|
|
+};
|
|
|
+
|
|
|
+/** @addtogroup kdtrees_grp KD-tree classes and adaptors
|
|
|
+ * @{ */
|
|
|
+
|
|
|
+/** kd-tree static index
|
|
|
+ *
|
|
|
+ * Contains the k-d trees and other information for indexing a set of points
|
|
|
+ * for nearest-neighbor matching.
|
|
|
+ *
|
|
|
+ * The class "DatasetAdaptor" must provide the following interface (can be
|
|
|
+ * non-virtual, inlined methods):
|
|
|
+ *
|
|
|
+ * \code
|
|
|
+ * // Must return the number of data poins
|
|
|
+ * inline size_t kdtree_get_point_count() const { ... }
|
|
|
+ *
|
|
|
+ *
|
|
|
+ * // Must return the dim'th component of the idx'th point in the class:
|
|
|
+ * inline T kdtree_get_pt(const size_t idx, const size_t dim) const { ... }
|
|
|
+ *
|
|
|
+ * // Optional bounding-box computation: return false to default to a standard
|
|
|
+ * bbox computation loop.
|
|
|
+ * // Return true if the BBOX was already computed by the class and returned
|
|
|
+ * in "bb" so it can be avoided to redo it again.
|
|
|
+ * // Look at bb.size() to find out the expected dimensionality (e.g. 2 or 3
|
|
|
+ * for point clouds) template <class BBOX> bool kdtree_get_bbox(BBOX &bb) const
|
|
|
+ * {
|
|
|
+ * bb[0].low = ...; bb[0].high = ...; // 0th dimension limits
|
|
|
+ * bb[1].low = ...; bb[1].high = ...; // 1st dimension limits
|
|
|
+ * ...
|
|
|
+ * return true;
|
|
|
+ * }
|
|
|
+ *
|
|
|
+ * \endcode
|
|
|
+ *
|
|
|
+ * \tparam DatasetAdaptor The user-provided adaptor (see comments above).
|
|
|
+ * \tparam Distance The distance metric to use: nanoflann::metric_L1,
|
|
|
+ * nanoflann::metric_L2, nanoflann::metric_L2_Simple, etc. \tparam DIM
|
|
|
+ * Dimensionality of data points (e.g. 3 for 3D points) \tparam IndexType Will
|
|
|
+ * be typically size_t or int
|
|
|
+ */
|
|
|
+template <typename Distance, class DatasetAdaptor, int DIM = -1,
|
|
|
+ typename IndexType = size_t>
|
|
|
+class KDTreeSingleIndexAdaptor
|
|
|
+ : public KDTreeBaseClass<
|
|
|
+ KDTreeSingleIndexAdaptor<Distance, DatasetAdaptor, DIM, IndexType>,
|
|
|
+ Distance, DatasetAdaptor, DIM, IndexType> {
|
|
|
+public:
|
|
|
+ /** Deleted copy constructor*/
|
|
|
+ KDTreeSingleIndexAdaptor(
|
|
|
+ const KDTreeSingleIndexAdaptor<Distance, DatasetAdaptor, DIM, IndexType>
|
|
|
+ &) = delete;
|
|
|
+
|
|
|
+ /**
|
|
|
+ * The dataset used by this index
|
|
|
+ */
|
|
|
+ const DatasetAdaptor &dataset; //!< The source of our data
|
|
|
+
|
|
|
+ const KDTreeSingleIndexAdaptorParams index_params;
|
|
|
+
|
|
|
+ Distance distance;
|
|
|
+
|
|
|
+ typedef typename nanoflann::KDTreeBaseClass<
|
|
|
+ nanoflann::KDTreeSingleIndexAdaptor<Distance, DatasetAdaptor, DIM,
|
|
|
+ IndexType>,
|
|
|
+ Distance, DatasetAdaptor, DIM, IndexType>
|
|
|
+ BaseClassRef;
|
|
|
+
|
|
|
+ typedef typename BaseClassRef::ElementType ElementType;
|
|
|
+ typedef typename BaseClassRef::DistanceType DistanceType;
|
|
|
+
|
|
|
+ typedef typename BaseClassRef::Node Node;
|
|
|
+ typedef Node *NodePtr;
|
|
|
+
|
|
|
+ typedef typename BaseClassRef::Interval Interval;
|
|
|
+ /** Define "BoundingBox" as a fixed-size or variable-size container depending
|
|
|
+ * on "DIM" */
|
|
|
+ typedef typename BaseClassRef::BoundingBox BoundingBox;
|
|
|
+
|
|
|
+ /** Define "distance_vector_t" as a fixed-size or variable-size container
|
|
|
+ * depending on "DIM" */
|
|
|
+ typedef typename BaseClassRef::distance_vector_t distance_vector_t;
|
|
|
+
|
|
|
+ /**
|
|
|
+ * KDTree constructor
|
|
|
+ *
|
|
|
+ * Refer to docs in README.md or online in
|
|
|
+ * https://github.com/jlblancoc/nanoflann
|
|
|
+ *
|
|
|
+ * The KD-Tree point dimension (the length of each point in the datase, e.g. 3
|
|
|
+ * for 3D points) is determined by means of:
|
|
|
+ * - The \a DIM template parameter if >0 (highest priority)
|
|
|
+ * - Otherwise, the \a dimensionality parameter of this constructor.
|
|
|
+ *
|
|
|
+ * @param inputData Dataset with the input features
|
|
|
+ * @param params Basically, the maximum leaf node size
|
|
|
+ */
|
|
|
+ KDTreeSingleIndexAdaptor(const int dimensionality,
|
|
|
+ const DatasetAdaptor &inputData,
|
|
|
+ const KDTreeSingleIndexAdaptorParams ¶ms =
|
|
|
+ KDTreeSingleIndexAdaptorParams())
|
|
|
+ : dataset(inputData), index_params(params), distance(inputData) {
|
|
|
+ BaseClassRef::root_node = NULL;
|
|
|
+ BaseClassRef::m_size = dataset.kdtree_get_point_count();
|
|
|
+ BaseClassRef::m_size_at_index_build = BaseClassRef::m_size;
|
|
|
+ BaseClassRef::dim = dimensionality;
|
|
|
+ if (DIM > 0)
|
|
|
+ BaseClassRef::dim = DIM;
|
|
|
+ BaseClassRef::m_leaf_max_size = params.leaf_max_size;
|
|
|
+
|
|
|
+ // Create a permutable array of indices to the input vectors.
|
|
|
+ init_vind();
|
|
|
+ }
|
|
|
+
|
|
|
+ /**
|
|
|
+ * Builds the index
|
|
|
+ */
|
|
|
+ void buildIndex() {
|
|
|
+ BaseClassRef::m_size = dataset.kdtree_get_point_count();
|
|
|
+ BaseClassRef::m_size_at_index_build = BaseClassRef::m_size;
|
|
|
+ init_vind();
|
|
|
+ this->freeIndex(*this);
|
|
|
+ BaseClassRef::m_size_at_index_build = BaseClassRef::m_size;
|
|
|
+ if (BaseClassRef::m_size == 0)
|
|
|
+ return;
|
|
|
+ computeBoundingBox(BaseClassRef::root_bbox);
|
|
|
+ BaseClassRef::root_node =
|
|
|
+ this->divideTree(*this, 0, BaseClassRef::m_size,
|
|
|
+ BaseClassRef::root_bbox); // construct the tree
|
|
|
+ }
|
|
|
+
|
|
|
+ /** \name Query methods
|
|
|
+ * @{ */
|
|
|
+
|
|
|
+ /**
|
|
|
+ * Find set of nearest neighbors to vec[0:dim-1]. Their indices are stored
|
|
|
+ * inside the result object.
|
|
|
+ *
|
|
|
+ * Params:
|
|
|
+ * result = the result object in which the indices of the
|
|
|
+ * nearest-neighbors are stored vec = the vector for which to search the
|
|
|
+ * nearest neighbors
|
|
|
+ *
|
|
|
+ * \tparam RESULTSET Should be any ResultSet<DistanceType>
|
|
|
+ * \return True if the requested neighbors could be found.
|
|
|
+ * \sa knnSearch, radiusSearch
|
|
|
+ */
|
|
|
+ template <typename RESULTSET>
|
|
|
+ bool findNeighbors(RESULTSET &result, const ElementType *vec,
|
|
|
+ const SearchParams &searchParams) const {
|
|
|
+ assert(vec);
|
|
|
+ if (this->size(*this) == 0)
|
|
|
+ return false;
|
|
|
+ if (!BaseClassRef::root_node)
|
|
|
+ throw std::runtime_error(
|
|
|
+ "[nanoflann] findNeighbors() called before building the index.");
|
|
|
+ float epsError = 1 + searchParams.eps;
|
|
|
+
|
|
|
+ distance_vector_t
|
|
|
+ dists; // fixed or variable-sized container (depending on DIM)
|
|
|
+ auto zero = static_cast<decltype(result.worstDist())>(0);
|
|
|
+ assign(dists, (DIM > 0 ? DIM : BaseClassRef::dim),
|
|
|
+ zero); // Fill it with zeros.
|
|
|
+ DistanceType distsq = this->computeInitialDistances(*this, vec, dists);
|
|
|
+ searchLevel(result, vec, BaseClassRef::root_node, distsq, dists,
|
|
|
+ epsError); // "count_leaf" parameter removed since was neither
|
|
|
+ // used nor returned to the user.
|
|
|
+ return result.full();
|
|
|
+ }
|
|
|
+
|
|
|
+ /**
|
|
|
+ * Find the "num_closest" nearest neighbors to the \a query_point[0:dim-1].
|
|
|
+ * Their indices are stored inside the result object. \sa radiusSearch,
|
|
|
+ * findNeighbors \note nChecks_IGNORED is ignored but kept for compatibility
|
|
|
+ * with the original FLANN interface. \return Number `N` of valid points in
|
|
|
+ * the result set. Only the first `N` entries in `out_indices` and
|
|
|
+ * `out_distances_sq` will be valid. Return may be less than `num_closest`
|
|
|
+ * only if the number of elements in the tree is less than `num_closest`.
|
|
|
+ */
|
|
|
+ size_t knnSearch(const ElementType *query_point, const size_t num_closest,
|
|
|
+ IndexType *out_indices, DistanceType *out_distances_sq,
|
|
|
+ const int /* nChecks_IGNORED */ = 10) const {
|
|
|
+ nanoflann::KNNResultSet<DistanceType, IndexType> resultSet(num_closest);
|
|
|
+ resultSet.init(out_indices, out_distances_sq);
|
|
|
+ this->findNeighbors(resultSet, query_point, nanoflann::SearchParams());
|
|
|
+ return resultSet.size();
|
|
|
+ }
|
|
|
+
|
|
|
+ /**
|
|
|
+ * Find all the neighbors to \a query_point[0:dim-1] within a maximum radius.
|
|
|
+ * The output is given as a vector of pairs, of which the first element is a
|
|
|
+ * point index and the second the corresponding distance. Previous contents of
|
|
|
+ * \a IndicesDists are cleared.
|
|
|
+ *
|
|
|
+ * If searchParams.sorted==true, the output list is sorted by ascending
|
|
|
+ * distances.
|
|
|
+ *
|
|
|
+ * For a better performance, it is advisable to do a .reserve() on the vector
|
|
|
+ * if you have any wild guess about the number of expected matches.
|
|
|
+ *
|
|
|
+ * \sa knnSearch, findNeighbors, radiusSearchCustomCallback
|
|
|
+ * \return The number of points within the given radius (i.e. indices.size()
|
|
|
+ * or dists.size() )
|
|
|
+ */
|
|
|
+ size_t
|
|
|
+ radiusSearch(const ElementType *query_point, const DistanceType &radius,
|
|
|
+ std::vector<std::pair<IndexType, DistanceType>> &IndicesDists,
|
|
|
+ const SearchParams &searchParams) const {
|
|
|
+ RadiusResultSet<DistanceType, IndexType> resultSet(radius, IndicesDists);
|
|
|
+ const size_t nFound =
|
|
|
+ radiusSearchCustomCallback(query_point, resultSet, searchParams);
|
|
|
+ if (searchParams.sorted)
|
|
|
+ std::sort(IndicesDists.begin(), IndicesDists.end(), IndexDist_Sorter());
|
|
|
+ return nFound;
|
|
|
+ }
|
|
|
+
|
|
|
+ /**
|
|
|
+ * Just like radiusSearch() but with a custom callback class for each point
|
|
|
+ * found in the radius of the query. See the source of RadiusResultSet<> as a
|
|
|
+ * start point for your own classes. \sa radiusSearch
|
|
|
+ */
|
|
|
+ template <class SEARCH_CALLBACK>
|
|
|
+ size_t radiusSearchCustomCallback(
|
|
|
+ const ElementType *query_point, SEARCH_CALLBACK &resultSet,
|
|
|
+ const SearchParams &searchParams = SearchParams()) const {
|
|
|
+ this->findNeighbors(resultSet, query_point, searchParams);
|
|
|
+ return resultSet.size();
|
|
|
+ }
|
|
|
+
|
|
|
+ /** @} */
|
|
|
+
|
|
|
+public:
|
|
|
+ /** Make sure the auxiliary list \a vind has the same size than the current
|
|
|
+ * dataset, and re-generate if size has changed. */
|
|
|
+ void init_vind() {
|
|
|
+ // Create a permutable array of indices to the input vectors.
|
|
|
+ BaseClassRef::m_size = dataset.kdtree_get_point_count();
|
|
|
+ if (BaseClassRef::vind.size() != BaseClassRef::m_size)
|
|
|
+ BaseClassRef::vind.resize(BaseClassRef::m_size);
|
|
|
+ for (size_t i = 0; i < BaseClassRef::m_size; i++)
|
|
|
+ BaseClassRef::vind[i] = i;
|
|
|
+ }
|
|
|
+
|
|
|
+ void computeBoundingBox(BoundingBox &bbox) {
|
|
|
+ resize(bbox, (DIM > 0 ? DIM : BaseClassRef::dim));
|
|
|
+ if (dataset.kdtree_get_bbox(bbox)) {
|
|
|
+ // Done! It was implemented in derived class
|
|
|
+ } else {
|
|
|
+ const size_t N = dataset.kdtree_get_point_count();
|
|
|
+ if (!N)
|
|
|
+ throw std::runtime_error("[nanoflann] computeBoundingBox() called but "
|
|
|
+ "no data points found.");
|
|
|
+ for (int i = 0; i < (DIM > 0 ? DIM : BaseClassRef::dim); ++i) {
|
|
|
+ bbox[i].low = bbox[i].high = this->dataset_get(*this, 0, i);
|
|
|
+ }
|
|
|
+ for (size_t k = 1; k < N; ++k) {
|
|
|
+ for (int i = 0; i < (DIM > 0 ? DIM : BaseClassRef::dim); ++i) {
|
|
|
+ if (this->dataset_get(*this, k, i) < bbox[i].low)
|
|
|
+ bbox[i].low = this->dataset_get(*this, k, i);
|
|
|
+ if (this->dataset_get(*this, k, i) > bbox[i].high)
|
|
|
+ bbox[i].high = this->dataset_get(*this, k, i);
|
|
|
+ }
|
|
|
+ }
|
|
|
+ }
|
|
|
+ }
|
|
|
+
|
|
|
+ /**
|
|
|
+ * Performs an exact search in the tree starting from a node.
|
|
|
+ * \tparam RESULTSET Should be any ResultSet<DistanceType>
|
|
|
+ * \return true if the search should be continued, false if the results are
|
|
|
+ * sufficient
|
|
|
+ */
|
|
|
+ template <class RESULTSET>
|
|
|
+ bool searchLevel(RESULTSET &result_set, const ElementType *vec,
|
|
|
+ const NodePtr node, DistanceType mindistsq,
|
|
|
+ distance_vector_t &dists, const float epsError) const {
|
|
|
+ /* If this is a leaf node, then do check and return. */
|
|
|
+ if ((node->child1 == NULL) && (node->child2 == NULL)) {
|
|
|
+ // count_leaf += (node->lr.right-node->lr.left); // Removed since was
|
|
|
+ // neither used nor returned to the user.
|
|
|
+ DistanceType worst_dist = result_set.worstDist();
|
|
|
+ for (IndexType i = node->node_type.lr.left; i < node->node_type.lr.right;
|
|
|
+ ++i) {
|
|
|
+ const IndexType index = BaseClassRef::vind[i]; // reorder... : i;
|
|
|
+ DistanceType dist = distance.evalMetric(
|
|
|
+ vec, index, (DIM > 0 ? DIM : BaseClassRef::dim));
|
|
|
+ if (dist < worst_dist) {
|
|
|
+ if (!result_set.addPoint(dist, BaseClassRef::vind[i])) {
|
|
|
+ // the resultset doesn't want to receive any more points, we're done
|
|
|
+ // searching!
|
|
|
+ return false;
|
|
|
+ }
|
|
|
+ }
|
|
|
+ }
|
|
|
+ return true;
|
|
|
+ }
|
|
|
+
|
|
|
+ /* Which child branch should be taken first? */
|
|
|
+ int idx = node->node_type.sub.divfeat;
|
|
|
+ ElementType val = vec[idx];
|
|
|
+ DistanceType diff1 = val - node->node_type.sub.divlow;
|
|
|
+ DistanceType diff2 = val - node->node_type.sub.divhigh;
|
|
|
+
|
|
|
+ NodePtr bestChild;
|
|
|
+ NodePtr otherChild;
|
|
|
+ DistanceType cut_dist;
|
|
|
+ if ((diff1 + diff2) < 0) {
|
|
|
+ bestChild = node->child1;
|
|
|
+ otherChild = node->child2;
|
|
|
+ cut_dist = distance.accum_dist(val, node->node_type.sub.divhigh, idx);
|
|
|
+ } else {
|
|
|
+ bestChild = node->child2;
|
|
|
+ otherChild = node->child1;
|
|
|
+ cut_dist = distance.accum_dist(val, node->node_type.sub.divlow, idx);
|
|
|
+ }
|
|
|
+
|
|
|
+ /* Call recursively to search next level down. */
|
|
|
+ if (!searchLevel(result_set, vec, bestChild, mindistsq, dists, epsError)) {
|
|
|
+ // the resultset doesn't want to receive any more points, we're done
|
|
|
+ // searching!
|
|
|
+ return false;
|
|
|
+ }
|
|
|
+
|
|
|
+ DistanceType dst = dists[idx];
|
|
|
+ mindistsq = mindistsq + cut_dist - dst;
|
|
|
+ dists[idx] = cut_dist;
|
|
|
+ if (mindistsq * epsError <= result_set.worstDist()) {
|
|
|
+ if (!searchLevel(result_set, vec, otherChild, mindistsq, dists,
|
|
|
+ epsError)) {
|
|
|
+ // the resultset doesn't want to receive any more points, we're done
|
|
|
+ // searching!
|
|
|
+ return false;
|
|
|
+ }
|
|
|
+ }
|
|
|
+ dists[idx] = dst;
|
|
|
+ return true;
|
|
|
+ }
|
|
|
+
|
|
|
+public:
|
|
|
+ /** Stores the index in a binary file.
|
|
|
+ * IMPORTANT NOTE: The set of data points is NOT stored in the file, so when
|
|
|
+ * loading the index object it must be constructed associated to the same
|
|
|
+ * source of data points used while building it. See the example:
|
|
|
+ * examples/saveload_example.cpp \sa loadIndex */
|
|
|
+ void saveIndex(FILE *stream) { this->saveIndex_(*this, stream); }
|
|
|
+
|
|
|
+ /** Loads a previous index from a binary file.
|
|
|
+ * IMPORTANT NOTE: The set of data points is NOT stored in the file, so the
|
|
|
+ * index object must be constructed associated to the same source of data
|
|
|
+ * points used while building the index. See the example:
|
|
|
+ * examples/saveload_example.cpp \sa loadIndex */
|
|
|
+ void loadIndex(FILE *stream) { this->loadIndex_(*this, stream); }
|
|
|
+
|
|
|
+}; // class KDTree
|
|
|
+
|
|
|
+/** kd-tree dynamic index
|
|
|
+ *
|
|
|
+ * Contains the k-d trees and other information for indexing a set of points
|
|
|
+ * for nearest-neighbor matching.
|
|
|
+ *
|
|
|
+ * The class "DatasetAdaptor" must provide the following interface (can be
|
|
|
+ * non-virtual, inlined methods):
|
|
|
+ *
|
|
|
+ * \code
|
|
|
+ * // Must return the number of data poins
|
|
|
+ * inline size_t kdtree_get_point_count() const { ... }
|
|
|
+ *
|
|
|
+ * // Must return the dim'th component of the idx'th point in the class:
|
|
|
+ * inline T kdtree_get_pt(const size_t idx, const size_t dim) const { ... }
|
|
|
+ *
|
|
|
+ * // Optional bounding-box computation: return false to default to a standard
|
|
|
+ * bbox computation loop.
|
|
|
+ * // Return true if the BBOX was already computed by the class and returned
|
|
|
+ * in "bb" so it can be avoided to redo it again.
|
|
|
+ * // Look at bb.size() to find out the expected dimensionality (e.g. 2 or 3
|
|
|
+ * for point clouds) template <class BBOX> bool kdtree_get_bbox(BBOX &bb) const
|
|
|
+ * {
|
|
|
+ * bb[0].low = ...; bb[0].high = ...; // 0th dimension limits
|
|
|
+ * bb[1].low = ...; bb[1].high = ...; // 1st dimension limits
|
|
|
+ * ...
|
|
|
+ * return true;
|
|
|
+ * }
|
|
|
+ *
|
|
|
+ * \endcode
|
|
|
+ *
|
|
|
+ * \tparam DatasetAdaptor The user-provided adaptor (see comments above).
|
|
|
+ * \tparam Distance The distance metric to use: nanoflann::metric_L1,
|
|
|
+ * nanoflann::metric_L2, nanoflann::metric_L2_Simple, etc. \tparam DIM
|
|
|
+ * Dimensionality of data points (e.g. 3 for 3D points) \tparam IndexType Will
|
|
|
+ * be typically size_t or int
|
|
|
+ */
|
|
|
+template <typename Distance, class DatasetAdaptor, int DIM = -1,
|
|
|
+ typename IndexType = size_t>
|
|
|
+class KDTreeSingleIndexDynamicAdaptor_
|
|
|
+ : public KDTreeBaseClass<KDTreeSingleIndexDynamicAdaptor_<
|
|
|
+ Distance, DatasetAdaptor, DIM, IndexType>,
|
|
|
+ Distance, DatasetAdaptor, DIM, IndexType> {
|
|
|
+public:
|
|
|
+ /**
|
|
|
+ * The dataset used by this index
|
|
|
+ */
|
|
|
+ const DatasetAdaptor &dataset; //!< The source of our data
|
|
|
+
|
|
|
+ KDTreeSingleIndexAdaptorParams index_params;
|
|
|
+
|
|
|
+ std::vector<int> &treeIndex;
|
|
|
+
|
|
|
+ Distance distance;
|
|
|
+
|
|
|
+ typedef typename nanoflann::KDTreeBaseClass<
|
|
|
+ nanoflann::KDTreeSingleIndexDynamicAdaptor_<Distance, DatasetAdaptor, DIM,
|
|
|
+ IndexType>,
|
|
|
+ Distance, DatasetAdaptor, DIM, IndexType>
|
|
|
+ BaseClassRef;
|
|
|
+
|
|
|
+ typedef typename BaseClassRef::ElementType ElementType;
|
|
|
+ typedef typename BaseClassRef::DistanceType DistanceType;
|
|
|
+
|
|
|
+ typedef typename BaseClassRef::Node Node;
|
|
|
+ typedef Node *NodePtr;
|
|
|
+
|
|
|
+ typedef typename BaseClassRef::Interval Interval;
|
|
|
+ /** Define "BoundingBox" as a fixed-size or variable-size container depending
|
|
|
+ * on "DIM" */
|
|
|
+ typedef typename BaseClassRef::BoundingBox BoundingBox;
|
|
|
+
|
|
|
+ /** Define "distance_vector_t" as a fixed-size or variable-size container
|
|
|
+ * depending on "DIM" */
|
|
|
+ typedef typename BaseClassRef::distance_vector_t distance_vector_t;
|
|
|
+
|
|
|
+ /**
|
|
|
+ * KDTree constructor
|
|
|
+ *
|
|
|
+ * Refer to docs in README.md or online in
|
|
|
+ * https://github.com/jlblancoc/nanoflann
|
|
|
+ *
|
|
|
+ * The KD-Tree point dimension (the length of each point in the datase, e.g. 3
|
|
|
+ * for 3D points) is determined by means of:
|
|
|
+ * - The \a DIM template parameter if >0 (highest priority)
|
|
|
+ * - Otherwise, the \a dimensionality parameter of this constructor.
|
|
|
+ *
|
|
|
+ * @param inputData Dataset with the input features
|
|
|
+ * @param params Basically, the maximum leaf node size
|
|
|
+ */
|
|
|
+ KDTreeSingleIndexDynamicAdaptor_(
|
|
|
+ const int dimensionality, const DatasetAdaptor &inputData,
|
|
|
+ std::vector<int> &treeIndex_,
|
|
|
+ const KDTreeSingleIndexAdaptorParams ¶ms =
|
|
|
+ KDTreeSingleIndexAdaptorParams())
|
|
|
+ : dataset(inputData), index_params(params), treeIndex(treeIndex_),
|
|
|
+ distance(inputData) {
|
|
|
+ BaseClassRef::root_node = NULL;
|
|
|
+ BaseClassRef::m_size = 0;
|
|
|
+ BaseClassRef::m_size_at_index_build = 0;
|
|
|
+ BaseClassRef::dim = dimensionality;
|
|
|
+ if (DIM > 0)
|
|
|
+ BaseClassRef::dim = DIM;
|
|
|
+ BaseClassRef::m_leaf_max_size = params.leaf_max_size;
|
|
|
+ }
|
|
|
+
|
|
|
+ /** Assignment operator definiton */
|
|
|
+ KDTreeSingleIndexDynamicAdaptor_
|
|
|
+ operator=(const KDTreeSingleIndexDynamicAdaptor_ &rhs) {
|
|
|
+ KDTreeSingleIndexDynamicAdaptor_ tmp(rhs);
|
|
|
+ std::swap(BaseClassRef::vind, tmp.BaseClassRef::vind);
|
|
|
+ std::swap(BaseClassRef::m_leaf_max_size, tmp.BaseClassRef::m_leaf_max_size);
|
|
|
+ std::swap(index_params, tmp.index_params);
|
|
|
+ std::swap(treeIndex, tmp.treeIndex);
|
|
|
+ std::swap(BaseClassRef::m_size, tmp.BaseClassRef::m_size);
|
|
|
+ std::swap(BaseClassRef::m_size_at_index_build,
|
|
|
+ tmp.BaseClassRef::m_size_at_index_build);
|
|
|
+ std::swap(BaseClassRef::root_node, tmp.BaseClassRef::root_node);
|
|
|
+ std::swap(BaseClassRef::root_bbox, tmp.BaseClassRef::root_bbox);
|
|
|
+ std::swap(BaseClassRef::pool, tmp.BaseClassRef::pool);
|
|
|
+ return *this;
|
|
|
+ }
|
|
|
+
|
|
|
+ /**
|
|
|
+ * Builds the index
|
|
|
+ */
|
|
|
+ void buildIndex() {
|
|
|
+ BaseClassRef::m_size = BaseClassRef::vind.size();
|
|
|
+ this->freeIndex(*this);
|
|
|
+ BaseClassRef::m_size_at_index_build = BaseClassRef::m_size;
|
|
|
+ if (BaseClassRef::m_size == 0)
|
|
|
+ return;
|
|
|
+ computeBoundingBox(BaseClassRef::root_bbox);
|
|
|
+ BaseClassRef::root_node =
|
|
|
+ this->divideTree(*this, 0, BaseClassRef::m_size,
|
|
|
+ BaseClassRef::root_bbox); // construct the tree
|
|
|
+ }
|
|
|
+
|
|
|
+ /** \name Query methods
|
|
|
+ * @{ */
|
|
|
+
|
|
|
+ /**
|
|
|
+ * Find set of nearest neighbors to vec[0:dim-1]. Their indices are stored
|
|
|
+ * inside the result object.
|
|
|
+ *
|
|
|
+ * Params:
|
|
|
+ * result = the result object in which the indices of the
|
|
|
+ * nearest-neighbors are stored vec = the vector for which to search the
|
|
|
+ * nearest neighbors
|
|
|
+ *
|
|
|
+ * \tparam RESULTSET Should be any ResultSet<DistanceType>
|
|
|
+ * \return True if the requested neighbors could be found.
|
|
|
+ * \sa knnSearch, radiusSearch
|
|
|
+ */
|
|
|
+ template <typename RESULTSET>
|
|
|
+ bool findNeighbors(RESULTSET &result, const ElementType *vec,
|
|
|
+ const SearchParams &searchParams) const {
|
|
|
+ assert(vec);
|
|
|
+ if (this->size(*this) == 0)
|
|
|
+ return false;
|
|
|
+ if (!BaseClassRef::root_node)
|
|
|
+ return false;
|
|
|
+ float epsError = 1 + searchParams.eps;
|
|
|
+
|
|
|
+ // fixed or variable-sized container (depending on DIM)
|
|
|
+ distance_vector_t dists;
|
|
|
+ // Fill it with zeros.
|
|
|
+ assign(dists, (DIM > 0 ? DIM : BaseClassRef::dim),
|
|
|
+ static_cast<typename distance_vector_t::value_type>(0));
|
|
|
+ DistanceType distsq = this->computeInitialDistances(*this, vec, dists);
|
|
|
+ searchLevel(result, vec, BaseClassRef::root_node, distsq, dists,
|
|
|
+ epsError); // "count_leaf" parameter removed since was neither
|
|
|
+ // used nor returned to the user.
|
|
|
+ return result.full();
|
|
|
+ }
|
|
|
+
|
|
|
+ /**
|
|
|
+ * Find the "num_closest" nearest neighbors to the \a query_point[0:dim-1].
|
|
|
+ * Their indices are stored inside the result object. \sa radiusSearch,
|
|
|
+ * findNeighbors \note nChecks_IGNORED is ignored but kept for compatibility
|
|
|
+ * with the original FLANN interface. \return Number `N` of valid points in
|
|
|
+ * the result set. Only the first `N` entries in `out_indices` and
|
|
|
+ * `out_distances_sq` will be valid. Return may be less than `num_closest`
|
|
|
+ * only if the number of elements in the tree is less than `num_closest`.
|
|
|
+ */
|
|
|
+ size_t knnSearch(const ElementType *query_point, const size_t num_closest,
|
|
|
+ IndexType *out_indices, DistanceType *out_distances_sq,
|
|
|
+ const int /* nChecks_IGNORED */ = 10) const {
|
|
|
+ nanoflann::KNNResultSet<DistanceType, IndexType> resultSet(num_closest);
|
|
|
+ resultSet.init(out_indices, out_distances_sq);
|
|
|
+ this->findNeighbors(resultSet, query_point, nanoflann::SearchParams());
|
|
|
+ return resultSet.size();
|
|
|
+ }
|
|
|
+
|
|
|
+ /**
|
|
|
+ * Find all the neighbors to \a query_point[0:dim-1] within a maximum radius.
|
|
|
+ * The output is given as a vector of pairs, of which the first element is a
|
|
|
+ * point index and the second the corresponding distance. Previous contents of
|
|
|
+ * \a IndicesDists are cleared.
|
|
|
+ *
|
|
|
+ * If searchParams.sorted==true, the output list is sorted by ascending
|
|
|
+ * distances.
|
|
|
+ *
|
|
|
+ * For a better performance, it is advisable to do a .reserve() on the vector
|
|
|
+ * if you have any wild guess about the number of expected matches.
|
|
|
+ *
|
|
|
+ * \sa knnSearch, findNeighbors, radiusSearchCustomCallback
|
|
|
+ * \return The number of points within the given radius (i.e. indices.size()
|
|
|
+ * or dists.size() )
|
|
|
+ */
|
|
|
+ size_t
|
|
|
+ radiusSearch(const ElementType *query_point, const DistanceType &radius,
|
|
|
+ std::vector<std::pair<IndexType, DistanceType>> &IndicesDists,
|
|
|
+ const SearchParams &searchParams) const {
|
|
|
+ RadiusResultSet<DistanceType, IndexType> resultSet(radius, IndicesDists);
|
|
|
+ const size_t nFound =
|
|
|
+ radiusSearchCustomCallback(query_point, resultSet, searchParams);
|
|
|
+ if (searchParams.sorted)
|
|
|
+ std::sort(IndicesDists.begin(), IndicesDists.end(), IndexDist_Sorter());
|
|
|
+ return nFound;
|
|
|
+ }
|
|
|
+
|
|
|
+ /**
|
|
|
+ * Just like radiusSearch() but with a custom callback class for each point
|
|
|
+ * found in the radius of the query. See the source of RadiusResultSet<> as a
|
|
|
+ * start point for your own classes. \sa radiusSearch
|
|
|
+ */
|
|
|
+ template <class SEARCH_CALLBACK>
|
|
|
+ size_t radiusSearchCustomCallback(
|
|
|
+ const ElementType *query_point, SEARCH_CALLBACK &resultSet,
|
|
|
+ const SearchParams &searchParams = SearchParams()) const {
|
|
|
+ this->findNeighbors(resultSet, query_point, searchParams);
|
|
|
+ return resultSet.size();
|
|
|
+ }
|
|
|
+
|
|
|
+ /** @} */
|
|
|
+
|
|
|
+public:
|
|
|
+ void computeBoundingBox(BoundingBox &bbox) {
|
|
|
+ resize(bbox, (DIM > 0 ? DIM : BaseClassRef::dim));
|
|
|
+
|
|
|
+ if (dataset.kdtree_get_bbox(bbox)) {
|
|
|
+ // Done! It was implemented in derived class
|
|
|
+ } else {
|
|
|
+ const size_t N = BaseClassRef::m_size;
|
|
|
+ if (!N)
|
|
|
+ throw std::runtime_error("[nanoflann] computeBoundingBox() called but "
|
|
|
+ "no data points found.");
|
|
|
+ for (int i = 0; i < (DIM > 0 ? DIM : BaseClassRef::dim); ++i) {
|
|
|
+ bbox[i].low = bbox[i].high =
|
|
|
+ this->dataset_get(*this, BaseClassRef::vind[0], i);
|
|
|
+ }
|
|
|
+ for (size_t k = 1; k < N; ++k) {
|
|
|
+ for (int i = 0; i < (DIM > 0 ? DIM : BaseClassRef::dim); ++i) {
|
|
|
+ if (this->dataset_get(*this, BaseClassRef::vind[k], i) < bbox[i].low)
|
|
|
+ bbox[i].low = this->dataset_get(*this, BaseClassRef::vind[k], i);
|
|
|
+ if (this->dataset_get(*this, BaseClassRef::vind[k], i) > bbox[i].high)
|
|
|
+ bbox[i].high = this->dataset_get(*this, BaseClassRef::vind[k], i);
|
|
|
+ }
|
|
|
+ }
|
|
|
+ }
|
|
|
+ }
|
|
|
+
|
|
|
+ /**
|
|
|
+ * Performs an exact search in the tree starting from a node.
|
|
|
+ * \tparam RESULTSET Should be any ResultSet<DistanceType>
|
|
|
+ */
|
|
|
+ template <class RESULTSET>
|
|
|
+ void searchLevel(RESULTSET &result_set, const ElementType *vec,
|
|
|
+ const NodePtr node, DistanceType mindistsq,
|
|
|
+ distance_vector_t &dists, const float epsError) const {
|
|
|
+ /* If this is a leaf node, then do check and return. */
|
|
|
+ if ((node->child1 == NULL) && (node->child2 == NULL)) {
|
|
|
+ // count_leaf += (node->lr.right-node->lr.left); // Removed since was
|
|
|
+ // neither used nor returned to the user.
|
|
|
+ DistanceType worst_dist = result_set.worstDist();
|
|
|
+ for (IndexType i = node->node_type.lr.left; i < node->node_type.lr.right;
|
|
|
+ ++i) {
|
|
|
+ const IndexType index = BaseClassRef::vind[i]; // reorder... : i;
|
|
|
+ if (treeIndex[index] == -1)
|
|
|
+ continue;
|
|
|
+ DistanceType dist = distance.evalMetric(
|
|
|
+ vec, index, (DIM > 0 ? DIM : BaseClassRef::dim));
|
|
|
+ if (dist < worst_dist) {
|
|
|
+ if (!result_set.addPoint(
|
|
|
+ static_cast<typename RESULTSET::DistanceType>(dist),
|
|
|
+ static_cast<typename RESULTSET::IndexType>(
|
|
|
+ BaseClassRef::vind[i]))) {
|
|
|
+ // the resultset doesn't want to receive any more points, we're done
|
|
|
+ // searching!
|
|
|
+ return; // false;
|
|
|
+ }
|
|
|
+ }
|
|
|
+ }
|
|
|
+ return;
|
|
|
+ }
|
|
|
+
|
|
|
+ /* Which child branch should be taken first? */
|
|
|
+ int idx = node->node_type.sub.divfeat;
|
|
|
+ ElementType val = vec[idx];
|
|
|
+ DistanceType diff1 = val - node->node_type.sub.divlow;
|
|
|
+ DistanceType diff2 = val - node->node_type.sub.divhigh;
|
|
|
+
|
|
|
+ NodePtr bestChild;
|
|
|
+ NodePtr otherChild;
|
|
|
+ DistanceType cut_dist;
|
|
|
+ if ((diff1 + diff2) < 0) {
|
|
|
+ bestChild = node->child1;
|
|
|
+ otherChild = node->child2;
|
|
|
+ cut_dist = distance.accum_dist(val, node->node_type.sub.divhigh, idx);
|
|
|
+ } else {
|
|
|
+ bestChild = node->child2;
|
|
|
+ otherChild = node->child1;
|
|
|
+ cut_dist = distance.accum_dist(val, node->node_type.sub.divlow, idx);
|
|
|
+ }
|
|
|
+
|
|
|
+ /* Call recursively to search next level down. */
|
|
|
+ searchLevel(result_set, vec, bestChild, mindistsq, dists, epsError);
|
|
|
+
|
|
|
+ DistanceType dst = dists[idx];
|
|
|
+ mindistsq = mindistsq + cut_dist - dst;
|
|
|
+ dists[idx] = cut_dist;
|
|
|
+ if (mindistsq * epsError <= result_set.worstDist()) {
|
|
|
+ searchLevel(result_set, vec, otherChild, mindistsq, dists, epsError);
|
|
|
+ }
|
|
|
+ dists[idx] = dst;
|
|
|
+ }
|
|
|
+
|
|
|
+public:
|
|
|
+ /** Stores the index in a binary file.
|
|
|
+ * IMPORTANT NOTE: The set of data points is NOT stored in the file, so when
|
|
|
+ * loading the index object it must be constructed associated to the same
|
|
|
+ * source of data points used while building it. See the example:
|
|
|
+ * examples/saveload_example.cpp \sa loadIndex */
|
|
|
+ void saveIndex(FILE *stream) { this->saveIndex_(*this, stream); }
|
|
|
+
|
|
|
+ /** Loads a previous index from a binary file.
|
|
|
+ * IMPORTANT NOTE: The set of data points is NOT stored in the file, so the
|
|
|
+ * index object must be constructed associated to the same source of data
|
|
|
+ * points used while building the index. See the example:
|
|
|
+ * examples/saveload_example.cpp \sa loadIndex */
|
|
|
+ void loadIndex(FILE *stream) { this->loadIndex_(*this, stream); }
|
|
|
+};
|
|
|
+
|
|
|
+/** kd-tree dynaimic index
|
|
|
+ *
|
|
|
+ * class to create multiple static index and merge their results to behave as
|
|
|
+ * single dynamic index as proposed in Logarithmic Approach.
|
|
|
+ *
|
|
|
+ * Example of usage:
|
|
|
+ * examples/dynamic_pointcloud_example.cpp
|
|
|
+ *
|
|
|
+ * \tparam DatasetAdaptor The user-provided adaptor (see comments above).
|
|
|
+ * \tparam Distance The distance metric to use: nanoflann::metric_L1,
|
|
|
+ * nanoflann::metric_L2, nanoflann::metric_L2_Simple, etc. \tparam DIM
|
|
|
+ * Dimensionality of data points (e.g. 3 for 3D points) \tparam IndexType Will
|
|
|
+ * be typically size_t or int
|
|
|
+ */
|
|
|
+template <typename Distance, class DatasetAdaptor, int DIM = -1,
|
|
|
+ typename IndexType = size_t>
|
|
|
+class KDTreeSingleIndexDynamicAdaptor {
|
|
|
+public:
|
|
|
+ typedef typename Distance::ElementType ElementType;
|
|
|
+ typedef typename Distance::DistanceType DistanceType;
|
|
|
+
|
|
|
+protected:
|
|
|
+ size_t m_leaf_max_size;
|
|
|
+ size_t treeCount;
|
|
|
+ size_t pointCount;
|
|
|
+
|
|
|
+ /**
|
|
|
+ * The dataset used by this index
|
|
|
+ */
|
|
|
+ const DatasetAdaptor &dataset; //!< The source of our data
|
|
|
+
|
|
|
+ std::vector<int> treeIndex; //!< treeIndex[idx] is the index of tree in which
|
|
|
+ //!< point at idx is stored. treeIndex[idx]=-1
|
|
|
+ //!< means that point has been removed.
|
|
|
+
|
|
|
+ KDTreeSingleIndexAdaptorParams index_params;
|
|
|
+
|
|
|
+ int dim; //!< Dimensionality of each data point
|
|
|
+
|
|
|
+ typedef KDTreeSingleIndexDynamicAdaptor_<Distance, DatasetAdaptor, DIM>
|
|
|
+ index_container_t;
|
|
|
+ std::vector<index_container_t> index;
|
|
|
+
|
|
|
+public:
|
|
|
+ /** Get a const ref to the internal list of indices; the number of indices is
|
|
|
+ * adapted dynamically as the dataset grows in size. */
|
|
|
+ const std::vector<index_container_t> &getAllIndices() const { return index; }
|
|
|
+
|
|
|
+private:
|
|
|
+ /** finds position of least significant unset bit */
|
|
|
+ int First0Bit(IndexType num) {
|
|
|
+ int pos = 0;
|
|
|
+ while (num & 1) {
|
|
|
+ num = num >> 1;
|
|
|
+ pos++;
|
|
|
+ }
|
|
|
+ return pos;
|
|
|
+ }
|
|
|
+
|
|
|
+ /** Creates multiple empty trees to handle dynamic support */
|
|
|
+ void init() {
|
|
|
+ typedef KDTreeSingleIndexDynamicAdaptor_<Distance, DatasetAdaptor, DIM>
|
|
|
+ my_kd_tree_t;
|
|
|
+ std::vector<my_kd_tree_t> index_(
|
|
|
+ treeCount, my_kd_tree_t(dim /*dim*/, dataset, treeIndex, index_params));
|
|
|
+ index = index_;
|
|
|
+ }
|
|
|
+
|
|
|
+public:
|
|
|
+ Distance distance;
|
|
|
+
|
|
|
+ /**
|
|
|
+ * KDTree constructor
|
|
|
+ *
|
|
|
+ * Refer to docs in README.md or online in
|
|
|
+ * https://github.com/jlblancoc/nanoflann
|
|
|
+ *
|
|
|
+ * The KD-Tree point dimension (the length of each point in the datase, e.g. 3
|
|
|
+ * for 3D points) is determined by means of:
|
|
|
+ * - The \a DIM template parameter if >0 (highest priority)
|
|
|
+ * - Otherwise, the \a dimensionality parameter of this constructor.
|
|
|
+ *
|
|
|
+ * @param inputData Dataset with the input features
|
|
|
+ * @param params Basically, the maximum leaf node size
|
|
|
+ */
|
|
|
+ KDTreeSingleIndexDynamicAdaptor(const int dimensionality,
|
|
|
+ const DatasetAdaptor &inputData,
|
|
|
+ const KDTreeSingleIndexAdaptorParams ¶ms =
|
|
|
+ KDTreeSingleIndexAdaptorParams(),
|
|
|
+ const size_t maximumPointCount = 1000000000U)
|
|
|
+ : dataset(inputData), index_params(params), distance(inputData) {
|
|
|
+ treeCount = static_cast<size_t>(std::log2(maximumPointCount));
|
|
|
+ pointCount = 0U;
|
|
|
+ dim = dimensionality;
|
|
|
+ treeIndex.clear();
|
|
|
+ if (DIM > 0)
|
|
|
+ dim = DIM;
|
|
|
+ m_leaf_max_size = params.leaf_max_size;
|
|
|
+ init();
|
|
|
+ const size_t num_initial_points = dataset.kdtree_get_point_count();
|
|
|
+ if (num_initial_points > 0) {
|
|
|
+ addPoints(0, num_initial_points - 1);
|
|
|
+ }
|
|
|
+ }
|
|
|
+
|
|
|
+ /** Deleted copy constructor*/
|
|
|
+ KDTreeSingleIndexDynamicAdaptor(
|
|
|
+ const KDTreeSingleIndexDynamicAdaptor<Distance, DatasetAdaptor, DIM,
|
|
|
+ IndexType> &) = delete;
|
|
|
+
|
|
|
+ /** Add points to the set, Inserts all points from [start, end] */
|
|
|
+ void addPoints(IndexType start, IndexType end) {
|
|
|
+ size_t count = end - start + 1;
|
|
|
+ treeIndex.resize(treeIndex.size() + count);
|
|
|
+ for (IndexType idx = start; idx <= end; idx++) {
|
|
|
+ int pos = First0Bit(pointCount);
|
|
|
+ index[pos].vind.clear();
|
|
|
+ treeIndex[pointCount] = pos;
|
|
|
+ for (int i = 0; i < pos; i++) {
|
|
|
+ for (int j = 0; j < static_cast<int>(index[i].vind.size()); j++) {
|
|
|
+ index[pos].vind.push_back(index[i].vind[j]);
|
|
|
+ if (treeIndex[index[i].vind[j]] != -1)
|
|
|
+ treeIndex[index[i].vind[j]] = pos;
|
|
|
+ }
|
|
|
+ index[i].vind.clear();
|
|
|
+ index[i].freeIndex(index[i]);
|
|
|
+ }
|
|
|
+ index[pos].vind.push_back(idx);
|
|
|
+ index[pos].buildIndex();
|
|
|
+ pointCount++;
|
|
|
+ }
|
|
|
+ }
|
|
|
+
|
|
|
+ /** Remove a point from the set (Lazy Deletion) */
|
|
|
+ void removePoint(size_t idx) {
|
|
|
+ if (idx >= pointCount)
|
|
|
+ return;
|
|
|
+ treeIndex[idx] = -1;
|
|
|
+ }
|
|
|
+
|
|
|
+ /**
|
|
|
+ * Find set of nearest neighbors to vec[0:dim-1]. Their indices are stored
|
|
|
+ * inside the result object.
|
|
|
+ *
|
|
|
+ * Params:
|
|
|
+ * result = the result object in which the indices of the
|
|
|
+ * nearest-neighbors are stored vec = the vector for which to search the
|
|
|
+ * nearest neighbors
|
|
|
+ *
|
|
|
+ * \tparam RESULTSET Should be any ResultSet<DistanceType>
|
|
|
+ * \return True if the requested neighbors could be found.
|
|
|
+ * \sa knnSearch, radiusSearch
|
|
|
+ */
|
|
|
+ template <typename RESULTSET>
|
|
|
+ bool findNeighbors(RESULTSET &result, const ElementType *vec,
|
|
|
+ const SearchParams &searchParams) const {
|
|
|
+ for (size_t i = 0; i < treeCount; i++) {
|
|
|
+ index[i].findNeighbors(result, &vec[0], searchParams);
|
|
|
+ }
|
|
|
+ return result.full();
|
|
|
+ }
|
|
|
+};
|
|
|
+
|
|
|
+/** An L2-metric KD-tree adaptor for working with data directly stored in an
|
|
|
+ * Eigen Matrix, without duplicating the data storage. You can select whether a
|
|
|
+ * row or column in the matrix represents a point in the state space.
|
|
|
+ *
|
|
|
+ * Example of usage:
|
|
|
+ * \code
|
|
|
+ * Eigen::Matrix<num_t,Dynamic,Dynamic> mat;
|
|
|
+ * // Fill out "mat"...
|
|
|
+ *
|
|
|
+ * typedef KDTreeEigenMatrixAdaptor< Eigen::Matrix<num_t,Dynamic,Dynamic> >
|
|
|
+ * my_kd_tree_t; const int max_leaf = 10; my_kd_tree_t mat_index(mat, max_leaf
|
|
|
+ * ); mat_index.index->buildIndex(); mat_index.index->... \endcode
|
|
|
+ *
|
|
|
+ * \tparam DIM If set to >0, it specifies a compile-time fixed dimensionality
|
|
|
+ * for the points in the data set, allowing more compiler optimizations. \tparam
|
|
|
+ * Distance The distance metric to use: nanoflann::metric_L1,
|
|
|
+ * nanoflann::metric_L2, nanoflann::metric_L2_Simple, etc. \tparam row_major
|
|
|
+ * If set to true the rows of the matrix are used as the points, if set to false
|
|
|
+ * the columns of the matrix are used as the points.
|
|
|
+ */
|
|
|
+template <class MatrixType, int DIM = -1, class Distance = nanoflann::metric_L2,
|
|
|
+ bool row_major = true>
|
|
|
+struct KDTreeEigenMatrixAdaptor {
|
|
|
+ typedef KDTreeEigenMatrixAdaptor<MatrixType, DIM, Distance, row_major> self_t;
|
|
|
+ typedef typename MatrixType::Scalar num_t;
|
|
|
+ typedef typename MatrixType::Index IndexType;
|
|
|
+ typedef
|
|
|
+ typename Distance::template traits<num_t, self_t>::distance_t metric_t;
|
|
|
+ typedef KDTreeSingleIndexAdaptor<metric_t, self_t,
|
|
|
+ MatrixType::ColsAtCompileTime, IndexType>
|
|
|
+ index_t;
|
|
|
+
|
|
|
+ index_t *index; //! The kd-tree index for the user to call its methods as
|
|
|
+ //! usual with any other FLANN index.
|
|
|
+
|
|
|
+ /// Constructor: takes a const ref to the matrix object with the data points
|
|
|
+ KDTreeEigenMatrixAdaptor(const size_t dimensionality,
|
|
|
+ const std::reference_wrapper<const MatrixType> &mat,
|
|
|
+ const int leaf_max_size = 10)
|
|
|
+ : m_data_matrix(mat) {
|
|
|
+ const auto dims = row_major ? mat.get().cols() : mat.get().rows();
|
|
|
+ if (size_t(dims) != dimensionality)
|
|
|
+ throw std::runtime_error(
|
|
|
+ "Error: 'dimensionality' must match column count in data matrix");
|
|
|
+ if (DIM > 0 && int(dims) != DIM)
|
|
|
+ throw std::runtime_error(
|
|
|
+ "Data set dimensionality does not match the 'DIM' template argument");
|
|
|
+ index =
|
|
|
+ new index_t(static_cast<int>(dims), *this /* adaptor */,
|
|
|
+ nanoflann::KDTreeSingleIndexAdaptorParams(leaf_max_size));
|
|
|
+ index->buildIndex();
|
|
|
+ }
|
|
|
+
|
|
|
+public:
|
|
|
+ /** Deleted copy constructor */
|
|
|
+ KDTreeEigenMatrixAdaptor(const self_t &) = delete;
|
|
|
+
|
|
|
+ ~KDTreeEigenMatrixAdaptor() { delete index; }
|
|
|
+
|
|
|
+ const std::reference_wrapper<const MatrixType> m_data_matrix;
|
|
|
+
|
|
|
+ /** Query for the \a num_closest closest points to a given point (entered as
|
|
|
+ * query_point[0:dim-1]). Note that this is a short-cut method for
|
|
|
+ * index->findNeighbors(). The user can also call index->... methods as
|
|
|
+ * desired. \note nChecks_IGNORED is ignored but kept for compatibility with
|
|
|
+ * the original FLANN interface.
|
|
|
+ */
|
|
|
+ inline void query(const num_t *query_point, const size_t num_closest,
|
|
|
+ IndexType *out_indices, num_t *out_distances_sq,
|
|
|
+ const int /* nChecks_IGNORED */ = 10) const {
|
|
|
+ nanoflann::KNNResultSet<num_t, IndexType> resultSet(num_closest);
|
|
|
+ resultSet.init(out_indices, out_distances_sq);
|
|
|
+ index->findNeighbors(resultSet, query_point, nanoflann::SearchParams());
|
|
|
+ }
|
|
|
+
|
|
|
+ /** @name Interface expected by KDTreeSingleIndexAdaptor
|
|
|
+ * @{ */
|
|
|
+
|
|
|
+ const self_t &derived() const { return *this; }
|
|
|
+ self_t &derived() { return *this; }
|
|
|
+
|
|
|
+ // Must return the number of data points
|
|
|
+ inline size_t kdtree_get_point_count() const {
|
|
|
+ if(row_major)
|
|
|
+ return m_data_matrix.get().rows();
|
|
|
+ else
|
|
|
+ return m_data_matrix.get().cols();
|
|
|
+ }
|
|
|
+
|
|
|
+ // Returns the dim'th component of the idx'th point in the class:
|
|
|
+ inline num_t kdtree_get_pt(const IndexType idx, size_t dim) const {
|
|
|
+ if(row_major)
|
|
|
+ return m_data_matrix.get().coeff(idx, IndexType(dim));
|
|
|
+ else
|
|
|
+ return m_data_matrix.get().coeff(IndexType(dim), idx);
|
|
|
+ }
|
|
|
+
|
|
|
+ // Optional bounding-box computation: return false to default to a standard
|
|
|
+ // bbox computation loop.
|
|
|
+ // Return true if the BBOX was already computed by the class and returned in
|
|
|
+ // "bb" so it can be avoided to redo it again. Look at bb.size() to find out
|
|
|
+ // the expected dimensionality (e.g. 2 or 3 for point clouds)
|
|
|
+ template <class BBOX> bool kdtree_get_bbox(BBOX & /*bb*/) const {
|
|
|
+ return false;
|
|
|
+ }
|
|
|
+
|
|
|
+ /** @} */
|
|
|
+
|
|
|
+}; // end of KDTreeEigenMatrixAdaptor
|
|
|
+ /** @} */
|
|
|
+
|
|
|
+/** @} */ // end of grouping
|
|
|
+} // namespace nanoflann
|
|
|
+
|
|
|
+#endif /* NANOFLANN_HPP_ */
|